R statistics/rgrass
Installation
Terminology
See "Overview" in R_statistics
R within GRASS
In this example, we will use R within a GRASS GIS session, i.e. start R (or RStudio) from the GRASS GIS terminal or command line interface.
Startup
- First start a GRASS GIS session. Then, at the GRASS command line start R (for an 'rstudio' session, see below)
- In this example we will use the North Carolina sample dataset.
Reset the region settings to the defaults
GRASS (nc_spm_08_grass7):~ > g.region -d
Launch R from the GRASS prompt
GRASS (nc_spm_08_grass7):~ > R
Load the rgrass library:
library(rgrass)Get the GRASS environment (mapset, region, map projection, etc.); you can display the metadata for your location by printing G:
gmeta()
gisdbase /home/mneteler/grassdata
location nc_spm_08_grass7
mapset user1
rows 620
columns 1630
north 320000
south 10000
west 120000
east 935000
nsres 500
ewres 500
projection:
PROJCRS["NAD83(HARN) / North Carolina",
BASEGEOGCRS["NAD83(HARN)",
DATUM["NAD83 (High Accuracy Reference Network)",
ELLIPSOID["GRS 1980",6378137,298.257222101,
LENGTHUNIT["metre",1]]],
PRIMEM["Greenwich",0,
ANGLEUNIT["degree",0.0174532925199433]],
ID["EPSG",4152]],
CONVERSION["SPCS83 North Carolina zone (meters)",
METHOD["Lambert Conic Conformal (2SP)",
ID["EPSG",9802]],
PARAMETER["Latitude of false origin",33.75,
ANGLEUNIT["degree",0.0174532925199433],
ID["EPSG",8821]],
PARAMETER["Longitude of false origin",-79,
ANGLEUNIT["degree",0.0174532925199433],
ID["EPSG",8822]],
PARAMETER["Latitude of 1st standard parallel",36.1666666666667,
ANGLEUNIT["degree",0.0174532925199433],
ID["EPSG",8823]],
PARAMETER["Latitude of 2nd standard parallel",34.3333333333333,
ANGLEUNIT["degree",0.0174532925199433],
ID["EPSG",8824]],
PARAMETER["Easting at false origin",609601.22,
LENGTHUNIT["metre",1],
ID["EPSG",8826]],
PARAMETER["Northing at false origin",0,
LENGTHUNIT["metre",1],
ID["EPSG",8827]]],
CS[Cartesian,2],
AXIS["easting (X)",east,
ORDER[1],
LENGTHUNIT["metre",1]],
AXIS["northing (Y)",north,
ORDER[2],
LENGTHUNIT["metre",1]],
USAGE[
SCOPE["Engineering survey, topographic mapping."],
AREA["United States (USA) - North Carolina - counties of Alamance; Alexander; Alleghany; Anson; Ashe; Avery; Beaufort; Bertie; Bladen; Brunswick; Buncombe; Burke; Cabarrus; Caldwell; Camden; Carteret; Caswell; Catawba; Chatham; Cherokee; Chowan; Clay; Cleveland; Columbus; Craven; Cumberland; Currituck; Dare; Davidson; Davie; Duplin; Durham; Edgecombe; Forsyth; Franklin; Gaston; Gates; Graham; Granville; Greene; Guilford; Halifax; Harnett; Haywood; Henderson; Hertford; Hoke; Hyde; Iredell; Jackson; Johnston; Jones; Lee; Lenoir; Lincoln; Macon; Madison; Martin; McDowell; Mecklenburg; Mitchell; Montgomery; Moore; Nash; New Hanover; Northampton; Onslow; Orange; Pamlico; Pasquotank; Pender; Perquimans; Person; Pitt; Polk; Randolph; Richmond; Robeson; Rockingham; Rowan; Rutherford; Sampson; Scotland; Stanly; Stokes; Surry; Swain; Transylvania; Tyrrell; Union; Vance; Wake; Warren; Washington; Watauga; Wayne; Wilkes; Wilson; Yadkin; Yancey."],
BBOX[33.83,-84.33,36.59,-75.38]],
ID["EPSG",3358]]Listing of existing maps
List available vector maps:
execGRASS("g.list", parameters = list(type = "vector"))List selected vector maps (wildcard):
execGRASS("g.list", parameters = list(type = "vector", pattern = "precip*"))
precip
precip_30ynormals
precip_30ynormals_3dSave selected vector maps into R vector:
my_vmaps <- execGRASS("g.list", parameters = list(type = "vector", pattern = "precip*"))
attributes(my_vmaps)
attributes(my_vmaps)$resOut
[1] "precip" "precip_30ynormals" "precip_30ynormals_3d"List available raster maps:
execGRASS("g.list", parameters = list(type = "raster"))List selected raster maps (wildcard):
execGRASS("g.list", parameters = list(type = "raster", pattern = "lsat7_2002*"))
lsat7_2002_10
lsat7_2002_20
lsat7_2002_30
lsat7_2002_40
lsat7_2002_50
lsat7_2002_61
lsat7_2002_62
lsat7_2002_70
lsat7_2002_80Reading data from GRASS
Example 1: raster maps
Manual page: read_RAST()
Read two GRASS raster maps, "geology_30m" and "elevation", from the North Carolina sample data location into the R current session as a new object "ncdata". This object is a SpatRaster layer from terra package:
ncdata <- read_RAST(c("geology_30m", "elevation"))We can verify the new R object:
ncdata
class : SpatRaster
dimensions : 450, 500, 2 (nrow, ncol, nlyr)
resolution : 30, 30 (x, y)
extent : 630000, 645000, 215000, 228500 (xmin, xmax, ymin, ymax)
coord. ref. : +proj=lcc +lat_0=33.75 +lon_0=-79 +lat_1=36.1666666666667 +lat_2=34.3333333333333 +x_0=609601.22 +y_0=0 +ellps=GRS80 +units=m +no_defs
sources : file1da12a752d65.grd
file1da12112d9ea4.grd
names : geology_30m, elevation
min values : 217, 55.92321
max values : 948, 156.25197Now, let's plot one of the raster layers within the SpatRaster object. We'll load terra library to have all functionality available:
library(terra)
plot(ncdata$elevation, main="North Carolina elevation")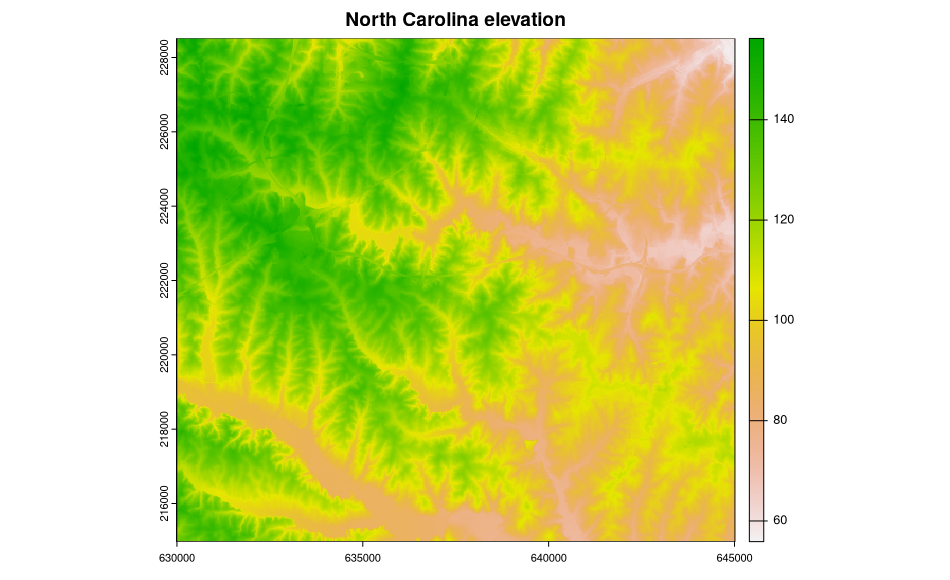
In addition, we can show what is going on inside the objects read into R:
summary(ncdata)
geology_30m elevation
Min. :217.0 Min. : 56.01
1st Qu.:217.0 1st Qu.: 94.77
Median :270.0 Median :108.86
Mean :311.2 Mean :110.36
3rd Qu.:270.0 3rd Qu.:126.80
Max. :948.0 Max. :156.07Example 2: single raster map
Here an example to transfer a single raster map from GRASS GIS to R:
library(rgrass)
library(terra)
execGRASS("g.region", raster = "elevation", flags = "p")
ncdata <- read_RAST("elevation")
summary(ncdata)
plot(ncdata)Example 3: vector maps
Here an example to transfer a single vector map from GRASS GIS to R. In this case, vector maps from GRASS will be converted into SpatVector objects from the terra package in R:
# needed prerequisite
library(rgrass)
library(terra)
execGRASS("v.info", map="schools_wake", layer="1")
vInfo("schools_wake")
myschools <- read_VECT("schools_wake")
print(summary(myschools))
plot(myschools)Example 4: Summarizing data
Create a box plot of geologic types at different elevations:
boxplot(ncdata$elevation ~ ncdata$geology_30m, medlwd = 1)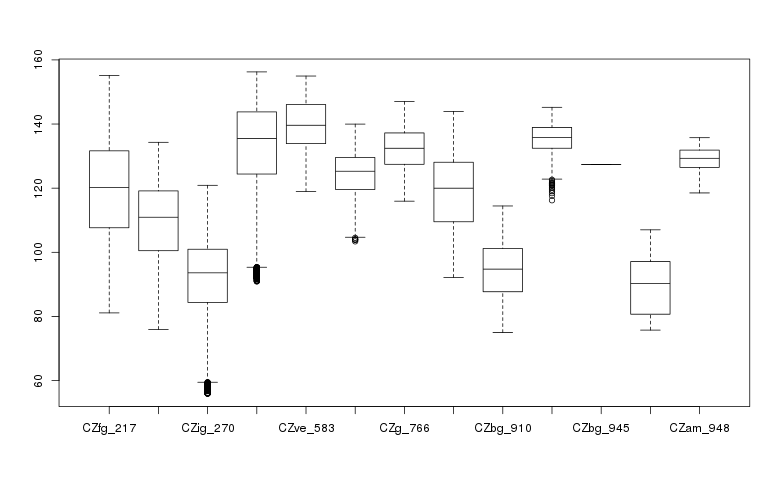
Example 5: Querying maps
Sometimes you may want to query GRASS GIS maps from your R session.
As an example, here the transfer of raster pixel values at the position of vector points:
# set the computational region first to the raster map:
execGRASS("g.region", raster = "elev_state_500m", flags = "p")
# query raster map at vector points, transfer result into R
goutput <- execGRASS("r.what", map="elev_state_500m", points="geodetic_pts", separator=",", intern=TRUE)
str(goutput)
chr [1:29939] "571530.81289275,265739.968425953,,187.8082200648" ...
# parse it
con <- textConnection(goutput)
go1 <- read.csv(con, header=FALSE)
str(go1)
'data.frame': 29939 obs. of 4 variables:
$ V1: num 571531 571359 571976 572391 573011 ...
$ V2: num 265740 265987 267049 267513 269615 ...
$ V3: logi NA NA NA NA NA NA ...
$ V4: Factor w/ 22738 levels "-0.0048115728",..: 6859 6642 6749 6411 6356 6904 7506 7224 6908 7167 ...
summary(go1)
V1 V2 V3 V4
Min. :121862 Min. : 7991 Mode:logical 0 : 723
1st Qu.:462706 1st Qu.:162508 NA's:29939 * : 293
Median :610328 Median :204502 0.3048 : 47
Mean :588514 Mean :200038 0.6096 : 44
3rd Qu.:717610 3rd Qu.:247633 1.524 : 42
Max. :946743 Max. :327186 0.9144 : 23
(Other):28767Exporting data back to GRASS
Finally, a SpatialGridDataFrame object is written back to a GRASS raster map:
First prepare some data: (square root of elevation)
ncdata$sqdem <- sqrt(ncdata$elevation)Export data from R back into a GRASS raster map:
writeRAST(ncdata, "sqdemNC", zcol="sqdem", ignore.stderr=TRUE)Check that it imported into GRASS ok:
execGRASS("r.info", parameters=list(map="sqdemNC"))+----------------------------------------------------------------------------+ | Map: sqdemNC Date: Sun Jul 19 13:06:34 2015 | | Mapset: PERMANENT Login of Creator: veroandreo | | Location: nc_spm_08_grass7 | | DataBase: /home/veroandreo/grassdata | | Title: ( sqdemNC ) | | Timestamp: none | |----------------------------------------------------------------------------| | | | Type of Map: raster Number of Categories: 0 | | Data Type: DCELL | | Rows: 491 | | Columns: 400 | | Total Cells: 196400 | | Projection: Lambert Conformal Conic | | N: 228500 S: 215000.0002 Res: 27.49490794 | | E: 645000 W: 630000 Res: 37.5 | | Range of data: min = 7.47818253045085 max = 12.5000787351036 | | | | Data Description: | | generated by r.in.bin | | | | Comments: | | | +----------------------------------------------------------------------------+
Using RStudio in a GRASS GIS session
If you are most used to run R through RStudio, but still want to have all GRASS data available for performing any analyses without loosing the possibility of still using GRASS command line, you can run:
GRASS> rstudio &
or, if you already are working on a certain RStudio project:
GRASS> rstudio /path/to/project/folder/ &
Then, you load rgrass library in your RStudio project
library(rgrass)and you are ready to go.
GRASS within R
Using GRASS GIS within a R session, i.e. you connect to a GRASS GIS location/mapset from within R (or RStudio).
Startup
In the first place, find out the path to the GRASS GIS library. This can be easily done with the following command (still outside of R; or through a system() call inside of R):
# OSGeo4W, Linux, Mac OSX users:
grass --config path
It may report something like:
# OSGeo4W users:
C:\OSGeo4W64\apps\grass\grass
# Linux, Mac OSX users:
/usr/local/grass
To call GRASS GIS functionality from R, start R and use the initGRASS() function to define the GRASS settings. If GRASS GIS was installed through OSGeo4W, first start the OSGeo4W Shell. Note that it may start in C: or a directory where the user does not have writing permission. Before calling R or RStudio, first change to a directory where you are sure you have writing permission.
## MS-Windows users (example for an OSGeo4W 64bit installation)
# change to a directory with writing permission
C:\>d:
D:\>cd temp
D:\temp>cd testR
# start R within this directory:
D:\temp\testR>R
# to start RStudio from OSGeo4W Shell, run: "C:/Program Files/RStudio/bin/rstudio.exe"
# load rgrass package
library(rgrass)
# initialisation of GRASS in the North Carolina sample dataset
initGRASS(gisBase = "C:/OSGeo4W64/apps/grass/grass",
gisDbase = "C:/Users/marissa/grassdata/",
location = "nc_spm_08_grass7",
mapset = "user1",
SG="elevation")Linux and Mac OSX users just need to start R or RStudio the usual way, load the rgrass package and initialize GRASS GIS with initGRASS() as shown below:
# load rgrass package
library(rgrass)
# initialisation of GRASS in the North Carolina sample dataset
initGRASS(gisBase = "/usr/local/grass",
home = tempdir(),
gisDbase = "/home/veroandreo/grassdata/",
location = "nc_spm_08_grass7",
mapset = "user1",
SG="elevation")Note: the optional SG parameter is a 'SpatialGrid' object to define the ‘DEFAULT_WIND’ of the temporary location.
# set computational region to default (optional)
system("g.region -dp")
# verify metadata
gmeta()
# get two raster maps into R space
ncdata <- read_RAST(c("geology_30m", "elevation"), cat=c(TRUE, FALSE))
# calculate object summaries
summary(ncdata$geology_30m)
CZfg_217 CZlg_262 CZig_270 CZbg_405 CZve_583 CZam_720 CZg_766 CZam_862
292 78 277 102 8 1 2 25
CZbg_910 Km_921 CZam_946 NA's
18 5 2 1009790R in GRASS in batch mode
Run the following script with
R CMD BATCH batch.R
library(rgrass)
# initialisation of GRASS in the North Carolina sample dataset (example with a compiled version)
initGRASS(gisBase = "/home/veroandreo/software/grass/dist.x86_64-pc-linux-gnu",
home = tempdir(),
gisDbase = "/home/veroandreo/grassdata/",
location = "nc_spm_08_grass7", mapset = "user1", SG="elevation",
override = TRUE)
# set region to default
system("g.region -dp")
# verify
gmeta()
# read data into R
ncdata <- read_RAST(c("geology_30m", "elevation"), cat=c(TRUE, FALSE))
# summary of geology map
summary(ncdata$geology_30m)
proc.time()The result is (shorted here):
cat batch.Rout
R version 4.0.2 (2020-06-22) -- "Taking Off Again"
Copyright (C) 2020 The R Foundation for Statistical Computing
Platform: x86_64-redhat-linux-gnu (64-bit)
...
> library(rgrass)
Loading required package: XML
GRASS GIS interface loaded with GRASS version: (GRASS not running)
> # initialisation and the use of north carolina dataset
> initGRASS(gisBase = "/home/veroandreo/software/grass/dist.x86_64-pc-linux-gnu", home = tempdir(),
+ gisDbase = "/home/veroandreo/grassdata/",
+ location = "nc_spm_08_grass7", mapset = "user1", SG="elevation",
+ override = TRUE)
gisdbase /home/veroandreo/grassdata/
location nc_spm_08_grass7
mapset user1
rows 13500
columns 15000
north 228500
south 215000
west 630000
east 645000
nsres 1
ewres 1
projection +proj=lcc +lat_1=36.16666666666666 +lat_2=34.33333333333334
+lat_0=33.75 +lon_0=-79 +x_0=609601.22 +y_0=0 +no_defs +a=6378137
+rf=298.257222101 +towgs84=0.000,0.000,0.000 +type=crs +to_meter=1
> system("g.region -dp")
projection: 99 (Lambert Conformal Conic)
zone: 0
datum: nad83
ellipsoid: a=6378137 es=0.006694380022900787
north: 320000
south: 10000
west: 120000
east: 935000
nsres: 500
ewres: 500
rows: 620
cols: 1630
cells: 1010600
> gmeta()
gisdbase /home/veroandreo/grassdata/
location nc_spm_08_grass7
mapset user1
rows 620
columns 1630
north 320000
south 10000
...
> ncdata <- read_RAST(c("geology_30m", "elevation"), cat=c(TRUE, FALSE))
...
> summary(ncdata$geology_30m)
CZfg_217 CZlg_262 CZig_270 CZbg_405 CZve_583 CZam_720 CZg_766 CZam_862
292 78 277 102 8 1 2 25
CZbg_910 Km_921 CZam_946 NA's
18 5 2 1009790
> proc.time()
user system elapsed
8.061 2.016 10.048Troubleshooting
Running out of disk space
Linux: A common issue is that /tmp/ is used for temporary files albeit being often a small partition. To change that to a larger directory, you may add to your $HOME/.bashrc the entry:
# change TMP directory of R (note: of course also another directory than suggested here is fine)
mkdir -p $HOME/tmp
export TMP=$HOME/tmp
The drawback of this approach is that on modern Linux systems the /tmp/ is a fast RAM disk (based on tempfs) while HOME directories are often on slower spinning disks (unless you have a SSD drive). At least you no longer run out of disk space easily.