Lidar Analysis of Vegetation Structure: Difference between revisions
⚠️Dnewcomb (talk | contribs) No edit summary |
⚠️Wenzeslaus (talk | contribs) (add see also section) |
||
| (19 intermediate revisions by 2 users not shown) | |||
| Line 31: | Line 31: | ||
All data are located in c:\grasslidar\data, unless otherwise noted. | All data are located in c:\grasslidar\data, unless otherwise noted. | ||
Step 1: Creating GRASS Workspace | == Step 1: Creating GRASS Workspace == | ||
The first thing to do when starting to work in GRASS is to create a Location. GRASS Locations are single projection areas with a defined resolution and extent. The initial location can be easily created from an existing data set. | The first thing to do when starting to work in GRASS is to create a Location. GRASS Locations are single projection areas with a defined resolution and extent. The initial location can be easily created from an existing data set. | ||
| Line 99: | Line 99: | ||
[[File:Grasslidar_11.png|GRASS Startup Menu]]<br /> | [[File:Grasslidar_11.png|GRASS Startup Menu]]<br /> | ||
* Click on the PERMANENT Mapset | |||
* Click Start GRASS session | |||
You will then see the Layer Manager Window, the Map Display Window for Display 1. | |||
[[File:Grasslidar_12.png|GRASS Windows]]<br /> | |||
==Step 2: Adding a data layer == | |||
We are now ready to add data to the Mapset. We can either import Raster and Vector data using the GDAL translation library, or we can link to existing external data sets read only to reduce data duplication. In this case, we will link to the 1m elevation data we used to create the Location using the r.external command. | |||
* In the Layer Manager window, click File-->Link external data-->Link external raster data. | |||
[[File:Grasslidar_13.png|Link External Data Menu]]<br /> | |||
This brings up the r.external dialog. | |||
[[File:Grasslidar_14.png|Link External Data Menu 2]]<br /> | |||
We would like to treat the 4 tiles of LiDAR and elevation data as a single seamless unit, to accomplish this, we are going to first select the demo05ft.vrt image index as our input raster. | |||
* Browse to C:\grasslidar\data\dem05ft.vrt and select it. Note that the Add layers into layer tree is checked. For large data sets, leave this unchecked. | |||
* Click on the import settings tab at the bottom | |||
* Click on the box for Extend region extents based on new dataset. | |||
[[File:Grasslidar_15.png|Link External Data Menu 3]]<br /> | |||
* Click back to the Source settings tab and Click Link. | |||
The Layer Manager window will switch to the Command console tab and display the results of the r.external command . Click the close button on the r.external dialog. | |||
[[File:Grasslidar_16.png|DEM View]]<br /> | |||
The 4 DEM rasters are displayed in the Map Display window as a single seamless data set . Any GIS software built with Geodata Abstraction Library(GDAL) ( including ArcGIS 10.0 and above) will see a vrt mosaic as a single raster layer. Creating the vrt is as simple as going to the black GRASS command window ,navigating to the directory in the with the rasters and typing: gdalbuildvrt dem05ft.vrt *.tif | |||
For larger data sets, it takes much less time to link to a raster data set than import it and linking also saves on storage space. | |||
== Step 3: Setting up the region == | |||
The region in a GRASS mapset defines the extent and resolution of the results of any raster actions. To make sure that all of a raster layer is affected by a command, it is customary to set the region’s extent and resolution to match the raster layer. To set the region to match the elevation raster that we just linked to , | |||
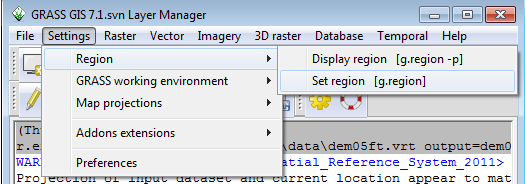
* From the Layer Manager window drop down options, click the Settings-->Region-->Set region | |||
[[File:Grasslidar_17.png|DEM View]]<br /> | |||
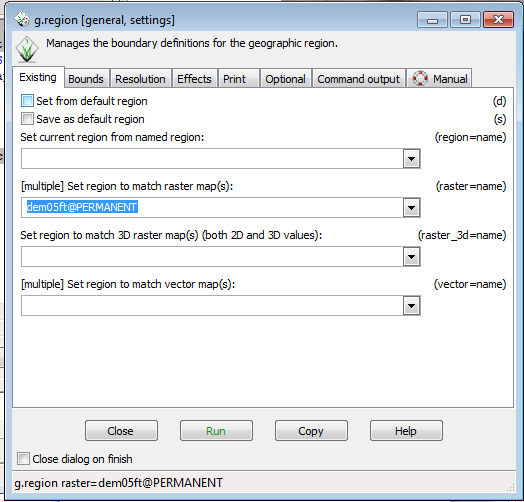
* In the g.region menu, in the Set region to match raster map section, use the drop down to select dem05ft@Permanent. | |||
* Click Run | |||
[[File:Grasslidar_18.png|Region Settings Selection]]<br /> | |||
When the command finishes, your will see “Command finished” and the time taken in the g.region window . | |||
* Click Close in the g.region window | |||
* Click Settings-->Region-->Display region | |||
[[File:Grasslidar_19.png|Region Display Selection]]<br /> | |||
In the command console tab of the Layer Manager window, you see the current region definintion: resolution, bounds, rows and columns. The output resolution of raster layers created will usually match the region resolution. To change the output resolution, you will need to change the resolution of the region. | |||
[[File:Grasslidar_20.png|Extent and Resolution of Mapset]]<br /> | |||
A quick note about the results. The datum is not recognised and assumed to be WGS84 by GRASS. The datum is NAD83 (2011). The data coordinates have not changed, the datum information has not been recognized on import. We can work with the data and assign the correct datum information after export from GRASS. This should be corrected by the final release of this version of GRASS. | |||
== Step 4: Processing the Lidar data == | |||
There are 4 lidar tiles that correspond to the 4 tiles of elevation data. The data is in the Compressed LAS files (laz) format. LASZip, http://www.laszip.org/, is an open compression method that losslessly compresses LAS format LiDAR data files to about 25 -30 percent of the original size. | |||
[[File:Grasslidar_21.png|LAZ LiDAR files]]<br /> | |||
You will be using the GRASS 7.1 r.in lidar. R.in.lidar uses a binning process, performing an analysis on the points that fall in each raster cell. It’s best to process las tiles as a single unit to avoid border effects. There are two ways to do this: | |||
1) Merge the las files together into a single file using tools such as lastools lasmerge, https://rapidlasso.com/lastools/lasmerge/ . Many software packages are still limited to LAS format 1.2 which can only handle up to 4.2 billion points per file ( a current limitation in GRASS7.1) . This option can also take up hard drive space. | |||
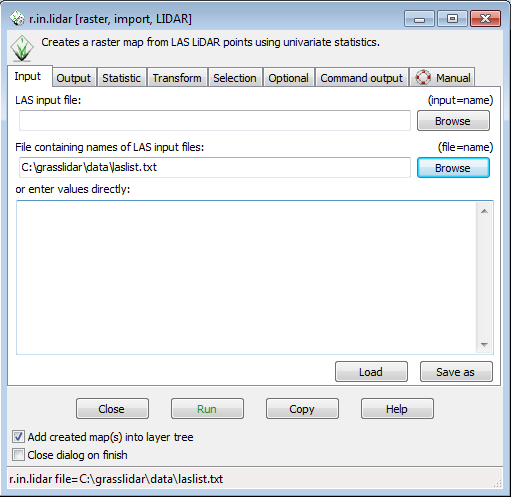
2) The second option available in r.in.lidar is to give a filename with a list of adjacent LAS/LAZ files and it will treat it as a virtual seamless LiDAR data set. If you look in the data directory, you will see laslist.txt. The contents of the file are: | |||
C:\grasslidar\data\LA_37_20026801_20141209.laz | |||
C:\grasslidar\data\LA_37_20026802_20141209.laz | |||
C:\grasslidar\data\LA_37_20026803_20141209.laz | |||
C:\grasslidar\data\LA_37_20026804_20141209.laz | |||
This is a list of the compressed las files in the directory that you will be processing. Give r.in.lidar c:\grasslidar\data\laslist.txt as the input file list, and it will treat all 4 files as a single point cloud. This method has been tested on file lists up to 966 las files. | |||
This exercise will use the second method. To start r.in.lidar, go to File-->Import raster data-->LAS Lidar points import. | |||
[[File:Grasslidar_22.png|Selecting r.in.lidar files]]<br /> | |||
This will bring up the r.in.lidar menu. | |||
You will notice that there are several tabs across the top of the command window. The first tab is the Input tab . You can either enter the name of the individual las or laz file, or you can enter the name of a txt file with a list of las or laz files. In the second option, browse to the location of the laslist.txt file and select it as shown. | |||
[[File:Grasslidar_23.png|r.in.lidar Input Tab]]<br /> | |||
Next we will set the output raster layer. | |||
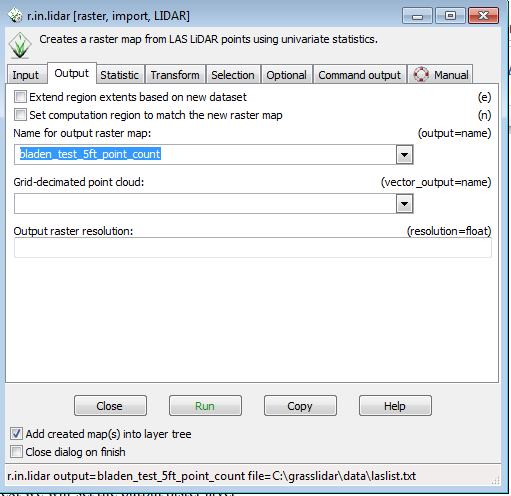
* Click on the Output tab | |||
* In the Name for output raster layer box, type “ bladen_test_5ft_pt_count” as the output layer | |||
[[File:Grasslidar_24.png|r.in.lidar Output Tab]]<br /> | |||
You will be binning the data using a 5ft resolution cell. In this case, you will be counting how many LiDAR returns are in each 5ft cell. | |||
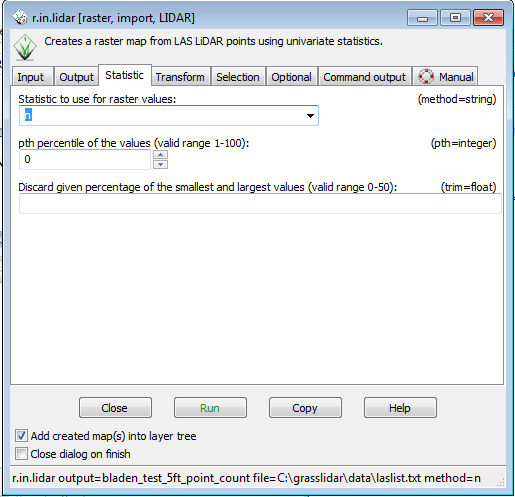
* Click on the Statistic tab | |||
* Select n from the Statistic to use for raster values drop down menu | |||
[[File:Grasslidar_25.png|r.in.lidar Statistic Tab]]<br /> | |||
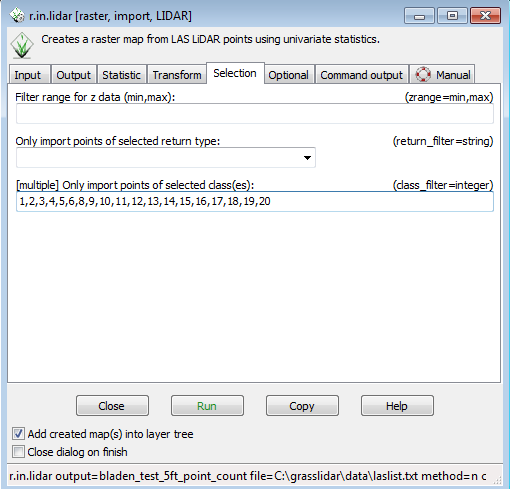
* Click on the selection tab | |||
* You want all points except the low noise ( class 7) so put 1,2,3,4,5,6,8,9,10,11,12,13,14,15,16,17,18,19,20 into the box for “Only import points of selected classes.” (feel free to copy and paste) | |||
[[File:Grasslidar_26.png|r.in.lidar Selection Tab]]<br /> | |||
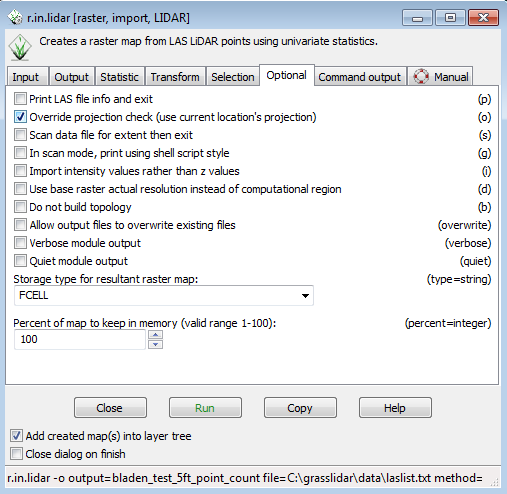
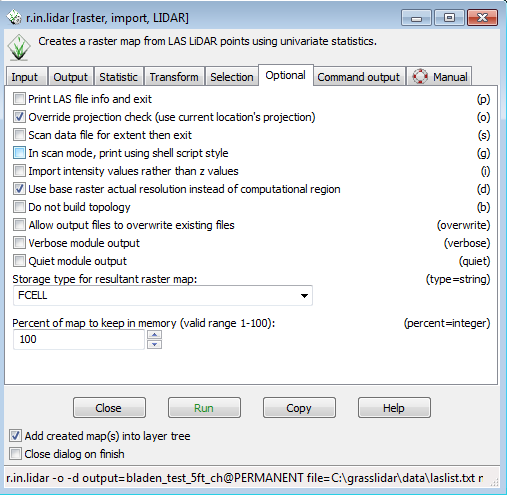
* Click on the Optional tab | |||
* Select the check box for Override projection check | |||
[[File:Grasslidar_27.png|r.in.lidar Options Tab]]<br /> | |||
* Click Run. | |||
Your display should look like this, with the result of the point count automatically added to the display. | |||
[[File:Grasslidar_28.png|r.in.lidar Count example Tab]]<br /> | |||
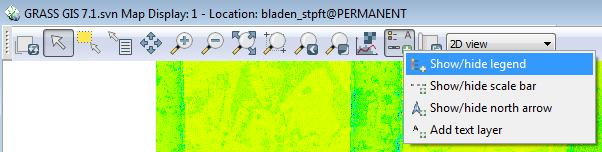
To add a legend to the display: | |||
* Click on the Add map elements button on the top of the Display window | |||
* Select show/hide legend | |||
[[File:Grasslidar_29.png|Legend example Tab]]<br /> | |||
The default legend is not clear for this data set, so simplify the legend by: | |||
* Double clicking on the legend to bring up the d.legend menu. | |||
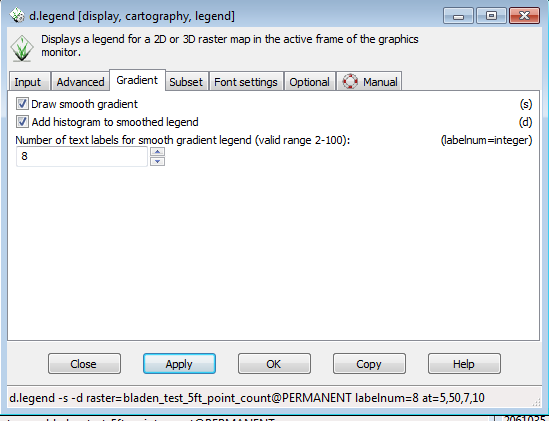
* Click on the Gradient tab and check the Draw smooth gradient and Add histogram to smoothed legend. | |||
* Change the number of text labels from the default of 5 to 8. | |||
* Click OK | |||
[[File:Grasslidar_30.png|Legend Gradient]]<br /> | |||
[[File:Grasslidar_31.png|Count display]]<br /> | |||
As you can see in the illustration above, the histogram on the legend tells you that most of the cells have fewer than 10 points per cell. For any valid statistical analysis of the point values in the cell you would need to pick a cell size larger than 5ft if you are performing statistics on the point values within each cell. | |||
You can also see the overlap of the LiDAR data collection flight lines. This makes the use of direct point count density useless as a measure of vegetation density unless all overlap points are removed. | |||
== Step 6: Canopy Heights and Other Forest Structure Metrics == | |||
Calculation of canopy heights involves determining the distance between the ground and the top of the vegetation. It can be done two steps , creating a digital surface model (DSM) and subtracting the Digital Elevation Model (DEM) With GRASS, this can be done in one step. | |||
Go back to the r.in.lidar menu: | |||
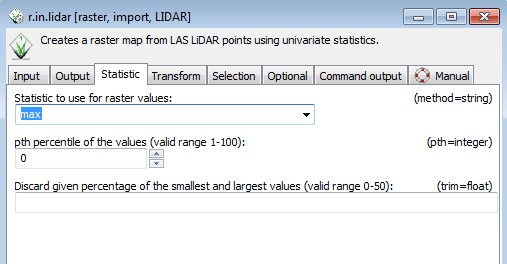
* Set the output raster to bladen_test_5ft_ch. | |||
* Go to the statistic tab and select max from the drop down menu. | |||
[[File:Grasslidar_32.png|Max Height]]<br /> | |||
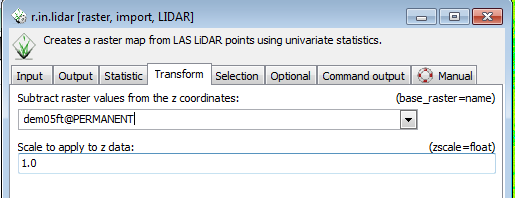
* Select the Transform tab | |||
* From the Subtract raster values from the z coordinates, select dem05ft from the dropdown menu. | |||
[[File:Grasslidar_33.png|r.inlidar Transform]]<br /> | |||
* Go to the Optional tab | |||
* Check the boxes next to Override projection check and Use base raster actual resolution instead of computational region | |||
[[File:Grasslidar_34.png|r.inlidar Options]]<br /> | |||
* Click Run | |||
* Select the Display window and double-click on the legend. | |||
* Select as the input raster layer bladen_test_05ft_ch | |||
You have just used the 5 ft resolution DEM to convert the z values of the point cloud to height above ground. Selecting the maximum value in each cell now gives the canopy height in each cell. Using this proceedure allows you to convert the LiDAR point Z values to height above ground at the 5ft resolution, but the calculations after the conversion will be done for the cell size of the current resolution of the location. | |||
Look back at the legend. You will notice that the bottom value is negative 4. Why is that? Run r.in.lidar again with the statistic st to min and the output raster set to bladen_test_05ft_min. | |||
* Go back to d.legend and set the raster to bladen_test_05ft_min. | |||
* Click on the Gradient tab and set the number of text labels to 20 | |||
* Click apply | |||
[[File:Grasslidar_35.png|Legend Options]]<br /> | |||
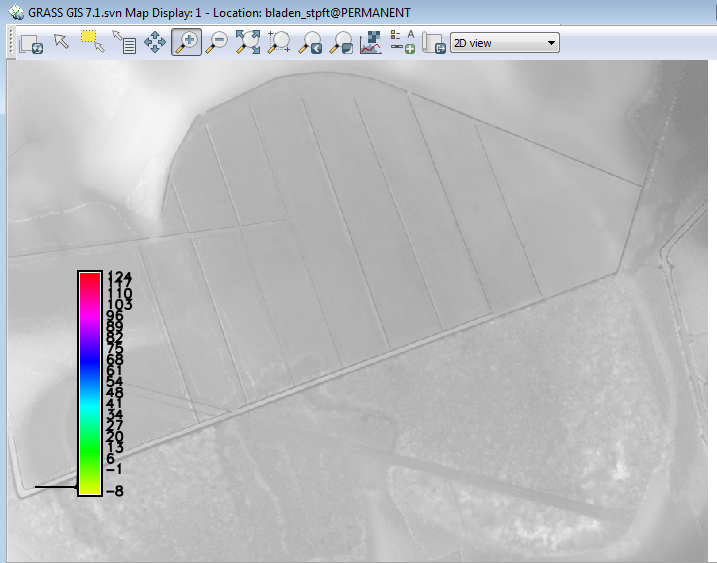
You can see that the bulk of the minimum values are less than the DEM surface. Turn off all layers except the DEM layer. Zoom into the large oval area in the Southeast corner of the DEM. | |||
[[File:Grasslidar_36.png|DEM ]]<br /> | |||
This feature is called a Carolina bay. The northern half has been ditched and drained, and the southern half remains in the native thick vegetation. Turn the min values back on. | |||
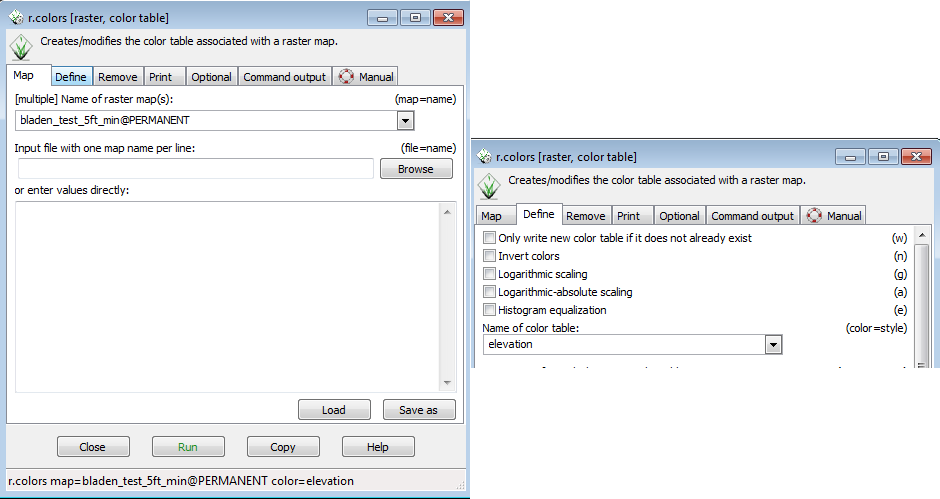
* Go to raster-->Manage colors --> Color tables. This will open the r.colors menu. | |||
* Select the Define tab and in the Name of color table dropdown select elevation. | |||
[[File:Grasslidar_38.png|Color editing ]]<br /> | |||
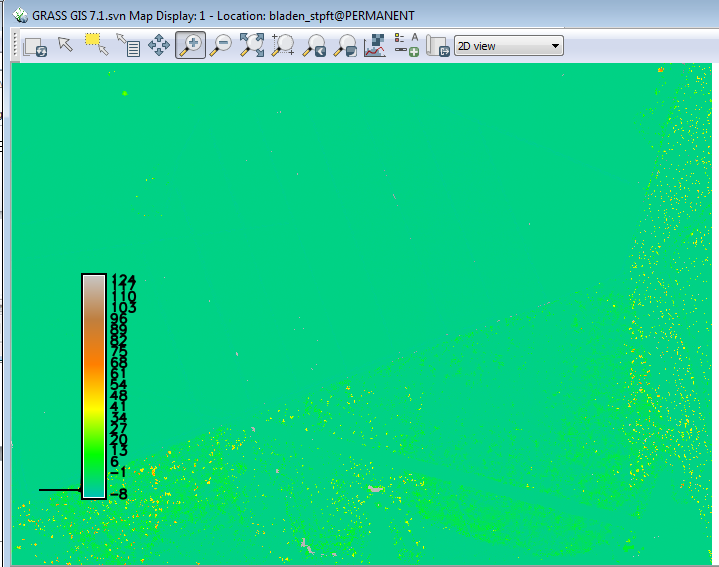
[[File:Grasslidar_39.png|Updated Colors ]]<br /> | |||
You will notice that in the light green and yellowish areas are significantly above the DEM surface. This is an indication that the laser did not penetrate the vegetation to hit the ground. While the DEM is not perfect, it gives us a better ground surface than just using the min value. There are ways to work around those differences, but that is not the topic for today. | |||
Look at the other end | |||
* Set the region to match the 5ft dem | |||
* Remove the base raster in the Transform tab and in the Optional tab | |||
* Remove the zrange on the Selection tab | |||
* Set the output raster to top_of_canopy_5ft | |||
* Run r.in.lidar with the max method | |||
Change your legend to match the top of canopy layer. What is the max height that you see? | |||
[[File:Grasslidar_40.png|Updated Colors Max Height ]]<br /> | |||
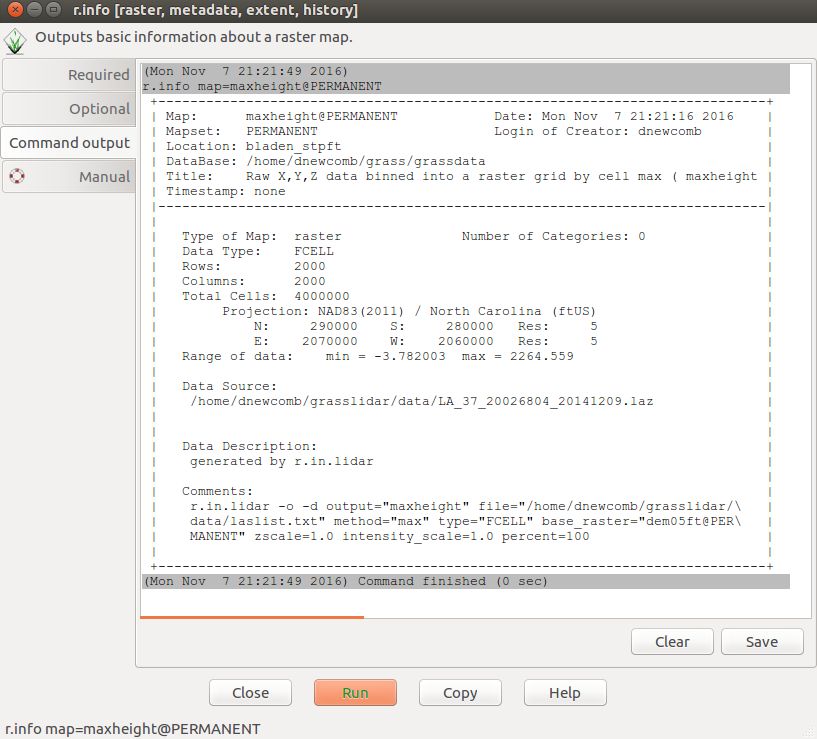
* Click Raster-->Reports and statistics-->Basic raster metadata (r.info) | |||
* Select dem05ft | |||
* Click Run | |||
[[File:Grasslidar_42.png| r.info output ]]<br /> | |||
R.info gives basic information about the raster layer. You see the rows, columns, number of cells, projection, the command that created the raster is listed as well. This is all useful information for metadata construction. The min and max values of the raster is what you need to look at now. The maximum value in the DEM is 2264.559. Since we have no trees in NC over 200ft, it seems we have some invalid high points. To remedy this:* | |||
* Go back to the Selection tab and and | |||
* for the zrange enter -10,250. | |||
* Click Run | |||
This gives a -10, 250 range around the high and low ground in the area (93-10 =83 and 128+250 = 378). This should exclude most of the extraneous points and leave a 5 ft resolution Digital Surface Model(DSM). Change the legend to display the new raster layer. What differences do you see? | |||
For now we are going to leave this dataset, but we will come back to it later. | |||
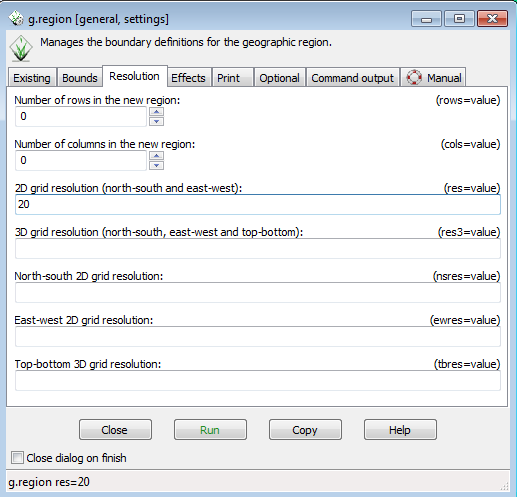
* Go back to Settings-->Region--> Set region to open the g.region dialog. | |||
* Select the Resolution tab. | |||
* Enter 20 in the 2D grid resolution box | |||
* Click Run | |||
[[File:Grasslidar_43.png| g.gregion menu ]]<br /> | |||
If you go to Settings-->Region-->Display region | |||
You will see : | |||
nsres: 20 | |||
ewres: 20 | |||
The Region resolution setting is now for 20ft cells. | |||
* Go back to the r.in.lidar menu and: | |||
* Set the output raster to bladen_test_20ft_ch and | |||
* Set the statistic to max. | |||
* Set the transform layer back to dem05ft | |||
* Check the box for use base raster resolution | |||
* Click Run | |||
* Set the legend to bladen_test_20ft_ch | |||
The Z values were converted to height above ground using the 5ft DEM and then statistics were calculated for 20ft cells. Notice how you see the outlines of the ditches. | |||
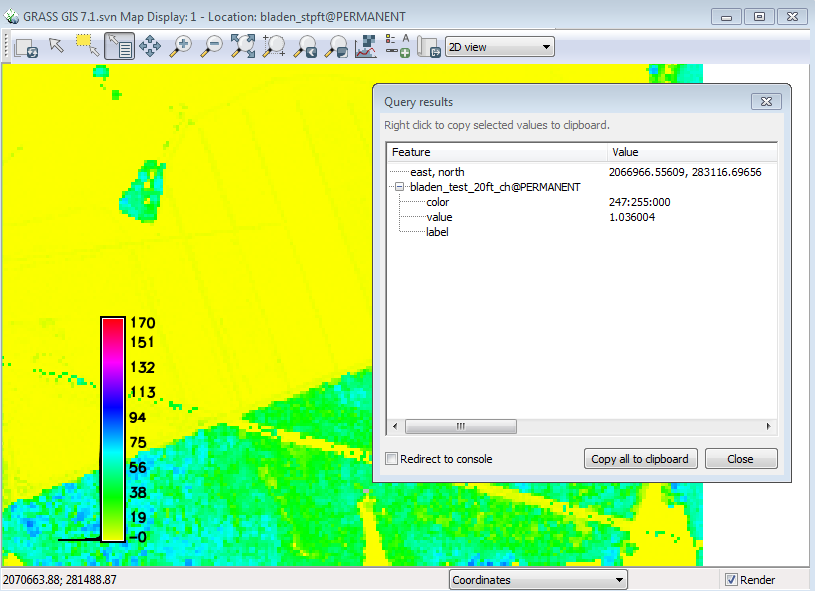
* Make sure that the bladen_test_20ft_ch is highlighted | |||
* Click on the Query selected raster/vector button ( 4th from the left in the display window) | |||
[[File:Grasslidar_44.png| Query button ]]<br /> | |||
* Place the crosshairs on one of the ditches | |||
* Left Click | |||
[[File:Grasslidar_45.png| Query result ]]<br /> | |||
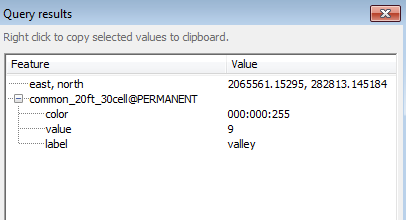
A window will pop up giving the location coordinates, the layer being queried, the color of the feature, and the value of the feature. In this case, the canopy height of the feature. The Z conversion to height above ground is not perfect and may depend on the accuracy of the DEM and the steepness of the slope represented by the raster. | |||
Remember that a slope is a smooth surface that is being interpolated to a grid format. | |||
Choosing a cell size can be important. At a 20ft resolution, you will be getting parts of larger trees and 60ft may be more appropriate to capture a more completepicture of the landscape vegetaion. Converting to height above ground allows for the use of cells of arbitrary size. Your analysis can match up with the cell size and footprint to exactly match the cell alignment of existing data. | |||
There are several other statistical measures that are available in r.in lidar: n, mean, range, sum, standard deviation, variance, coefficeint of variance, median,percentile, skewness, trimmean. Mean height, variance and skewness are metrics used in vegetation analysis. | |||
At the 20ft resolution, repeat r.in.lidar for variance and mean methods. What differences do you see in the resulting rasters? | |||
Repeat r.in.lidar at 20ft resolution using n as the statistic to create a point count layer. | |||
* Go to the Selection tab and | |||
* Set the z range to 3,10 | |||
* Set the output raster to bladen_20ft_shrub_count | |||
* Click Run | |||
* Go to the Selection tab | |||
* Set the zrange to 10,20 | |||
* Set the output raster to bladen_20ft_midstory_count | |||
* Click Run | |||
As stated above, using the raw point count is not suitable for vegetation analysis in areas where there are overlaps in collection. However, if horizontal sections of the point counts per cell are divided by the total point count per cell you can compare the percentage of the points that occur in horizontal sections. In this case, we will be using raster algebra to divide the shrub and midstory point counts by the total point counts using r.mapcalc, https://grass.osgeo.org/grass71/manuals/r.mapcalc.html . | |||
* Go to Raster-->Raster map calculator (r.mapcalc) | |||
[[File:Grasslidar_46.png| Raster Calculator Selection]]<br /> | |||
This will open the r.mapcalc menu: | |||
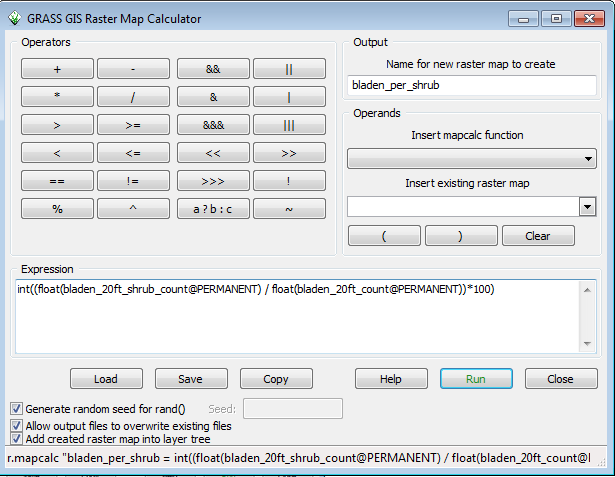
* Enter bladen_per_shrub in the Name for new raster map to create | |||
* Use the dropdown in the Insert existing raster map section to select bladen_20ft_shrub_count | |||
* Click on the / button for division | |||
* Use the dropdown to select bladen_20ft_count | |||
Now things get interesting. The point count rasters are integers. Mathmatical operations between integers results in an integer. Dividing an integer by a larger integer number results in a number less than one. In an integer grid, this truncates to 0. | |||
To get around this, you convert each integer grid before the operation to a floating point grid, so bladen_20ft_shrub becomes float(bladen_20ft_shrub) and bladen_20ft_count becomes float(bladen_20ft_count) and you divide one floating point grid by another floating point grid and you get a floating point grid. | |||
Multiply this floating point grid by 100 to put the values back in the range of 100 to 0. Finally, convert results of the entire operation back to an integer grid by enclosing it with int() . | |||
The final command in the Expression box is: | |||
int((float(bladen_20ft_shrub_count@PERMANENT)/float(bladen_20ft_count@PERMANENT))*100) | |||
[[File:Grasslidar_47.png| Raster Calculator Menu]]<br /> | |||
The resulting raster should look like this: | |||
[[File:Grasslidar_48.png| Raster Calculator Result]]<br /> | |||
Repeat the exercise with the midstory layer and compare the two layers. Do you see differences between the shrub and midstory percentage of points layers? | |||
We will continue the vegetation analysis, but first we need to take a short side trip into terrain analysis that will relate back to our vegetation analysis.. Terrain analysis will be done using geomrphons. | |||
== Step 5: Terrain analysis with geomorphons == | |||
What are geomorphons? Geomorphon is a new technique for classifying terrain based on a line of sight analysis, see http://geomorphometry.org/system/files/StepinskiJasiewicz2011geomorphometry.pdf , and http://www.sciencedirect.com/science/article/pii/S0169555X12005028 | |||
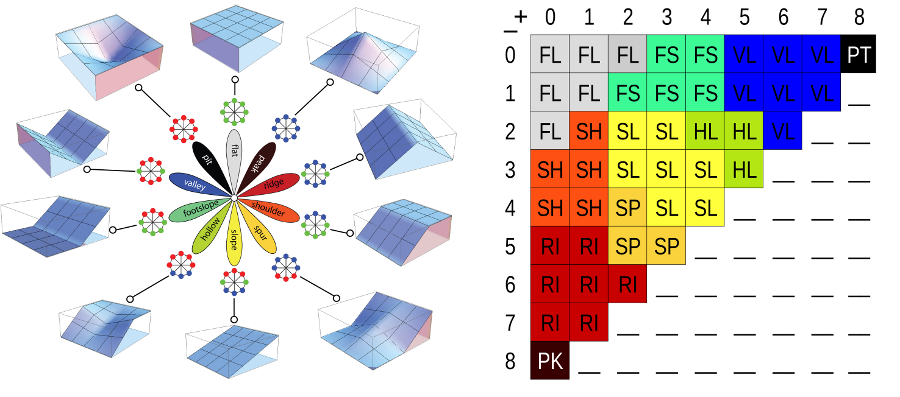
The technique uses a line of sight metric for terrain form calculation. There are options for a simple 10 class representation, a 498 class numeric coding, or positive and negative encoding per cell on line of sight. | |||
[[File:Grasslidar_49.png| Geomorphon Classes]]<br /> | |||
For this activity, we will use the simple 10 class calculation. Outputs will be cells with the following classes: Flat, peak, ridge, shoulder, spur, slope hollow, footslope, valley, and pit. | |||
To do our terrain classification, we will need to load the r.geomorphon addon to GRASS. | |||
GRASS has an online repository of Addons much like QGIS has for plugins. Most addons do not require administrative access to install. | |||
* In the Layer Manager window, click on Settings-->Addons extentions-->Install extention from addons | |||
[[File:Grasslidar_50.png| Addon Menu]]<br /> | |||
This will open the Fetch and install extension window. GRASS commands are named by their function: r.* commands are raster commands, v.* commands are vector commands, i.* commands are image analysis commands. | |||
* Click on the + next to raster to expand the view on all raster commands | |||
[[File:Grasslidar_51.png| Addon Selection]]<br /> | |||
* Select r.geomorphon | |||
* Click install | |||
In the Layer Manager window you can follow the progress of the installation. | |||
[[File:Grasslidar_52.png| Addon Installation]]<br /> | |||
In this case, downloading and installing the addon took 4 seconds on a 2010 vintage Windows 7 laptop. | |||
== Step 6: Running r.geomorphon == | |||
The r.geomorphon command documentation is at http://grass.osgeo.org/grass70/manuals/addons/r.geomorphon.html and the publications for the technique can be found at the end of the command description. | |||
To start r.geomorphon, | |||
In the GRASS Command window, type r.geomorphon and press Enter. | |||
[[File:Grasslidar_53.png| Start r.geomorphon]]<br /> | |||
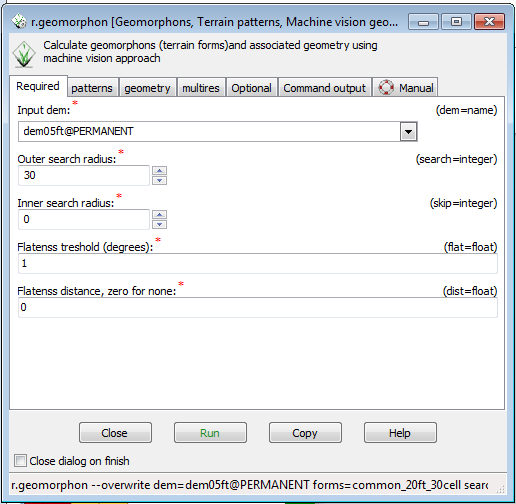
Entering the command without arguments brings up the r.geomorphon menu. You will notice that there are tabs across the top. The first tab is the Required Tab, with required data inputs to run the command. | |||
* From the dropdown for Input DEM, dem05ft@PERMANENT ( remember, the current resolution for the region is 20ft. GRASS will take the center value of the 5ft DEM in each 20ft cell and use that for the 20ft cell value, your output raster will be 20ft resolution) | |||
* In the Outer search radius, enter 30 ( number of cells around each cell to analyze) This is a very flat area, so the number may need to be higher to extract some features. | |||
[[File:Grasslidar_54.png| R.geomorphon setup]]<br /> | |||
You will note that as the command arguments are entered, they are echoed across the bottom. | |||
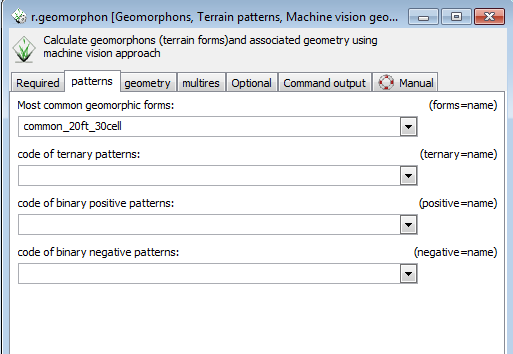
* Click on the patterns tab. | |||
* Type common_20ft_30cell in the box under Most common geomorphic forms. | |||
[[File:Grasslidar_55.png| R.geomorphon setup 2]]<br /> | |||
* Click Run | |||
The tab will immediately switch to the Command Output tab, the command options will be echoed , and a progress bar will show the progress of the command. | |||
You will need to manually add the result to the Display. | |||
* At the top of the Layer Manager window, Click Add raster map layer button | |||
[[File:Grasslidar_56.png| Select Raster Layer]]<br /> | |||
* In the d.rast menu that pops up, select common_20ft_30cell | |||
[[File:Grasslidar_57.png| Select Raster Layer d.rast]]<br /> | |||
* Click OK | |||
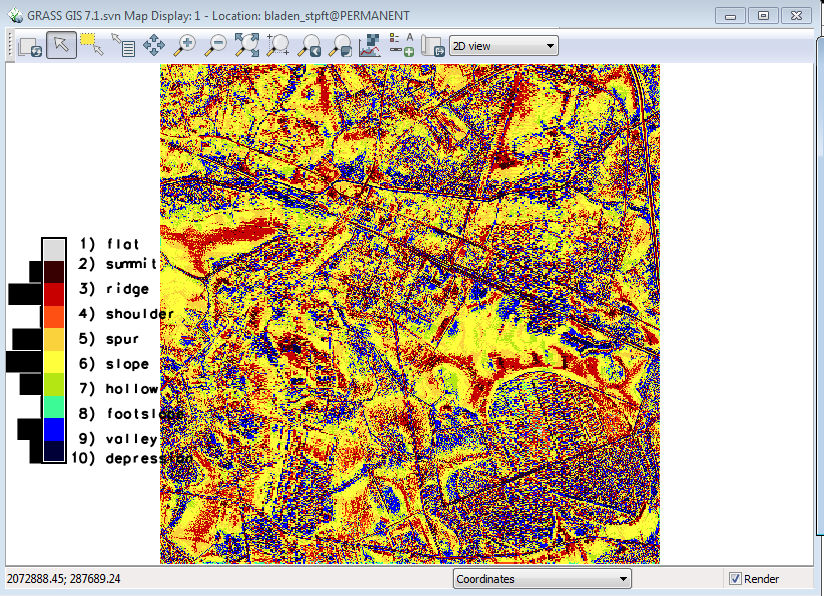
Your results should look like this ( once you select the legend to show the current raster): | |||
[[File:Grasslidar_58.png| Geomorphon Layer]]<br /> | |||
A check of the Windows Task Manager shows that the r.geomorphon command is using less than 100 MB of RAM to process the area. This process will take 7.5 minutes on a 2010 vintage i7 laptop. | |||
Larger areas and larger cell radius extents will require more memory. This process scales fairly well. It has been used on the Statewide 20ft elevation grid for North Carolina ( 6.6 billion cells) with a radius of 30 cells, but it required more than 4GB of RAM and took overnight to run on a Linux workstation. GRASS 7.2 on Windows is 64 bit and will address more than 4 GB of RAM on that platform. | |||
You can see a lot of different patterns, but what do they mean? Click on the Query raster / vector maps tool and you can click individual cells to extract the value. | |||
[[File:Grasslidar_59.png| Information Icon]]<br /> | |||
For example, a blue pixel represents integer value 9 or valley. | |||
[[File:Grasslidar_60.png| Query Results]]<br /> | |||
Alternatively, you can | |||
* Click on the Add map elements Button | |||
* Select Show/Hide legend | |||
* Click on the Gradient tab | |||
* Check the boxes for draw smooth Gradient and ad histogram | |||
* Put 10 in Number of text labels | |||
* Click Apply | |||
You should then see the following legend with the integer values, classes and histogram by class. | |||
[[File:Grasslidar_61.png| Geomorphon Legend]]<br /> | |||
Pretty handy way of describing the landscape. What does this have to do with vegetation? | |||
== Step 7: Extracting canopy features using geomorphons == | |||
* Add the bladen_test_5ft_ch ( add raster) . | |||
* Set the region to match bladen_test_5ft_ch(SettingsRegionSet regionSet region to match raster map) | |||
* Run a 3 cell common geomorphon analysis on the 5ft canopy height | |||
We want to look at the top of the canopy to pick out treetops and canopy openings. The treetops should be summits and the canopy openings should be depressions. We don’t care about any other class, so let’s create a new layer with just those two classes. | |||
* Go to Raster-->Change category values and classes-->Reclassify (r.reclass) | |||
* Select common_5ft_ch_3cell for the Name of raster to be reclassified. | |||
* Set the output raster to treetops_openings_5ft | |||
* Go down to the enter values directly box and type in: | |||
1 = Null | |||
2 = 1 Treetops | |||
3 thru 9 = NULL | |||
10 = 2 Openings | |||
* Turn off all other layers | |||
* Set your legend to treetops_openings_5ft | |||
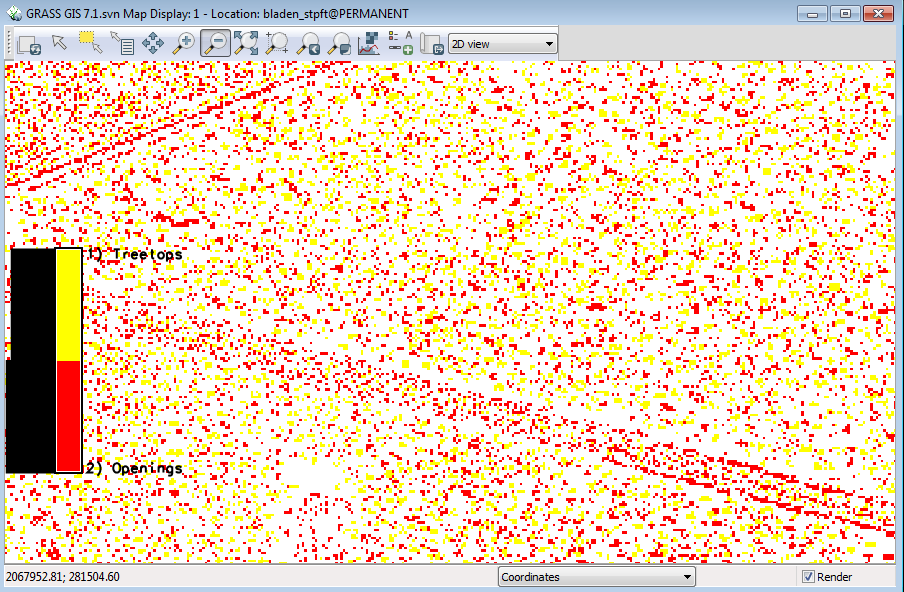
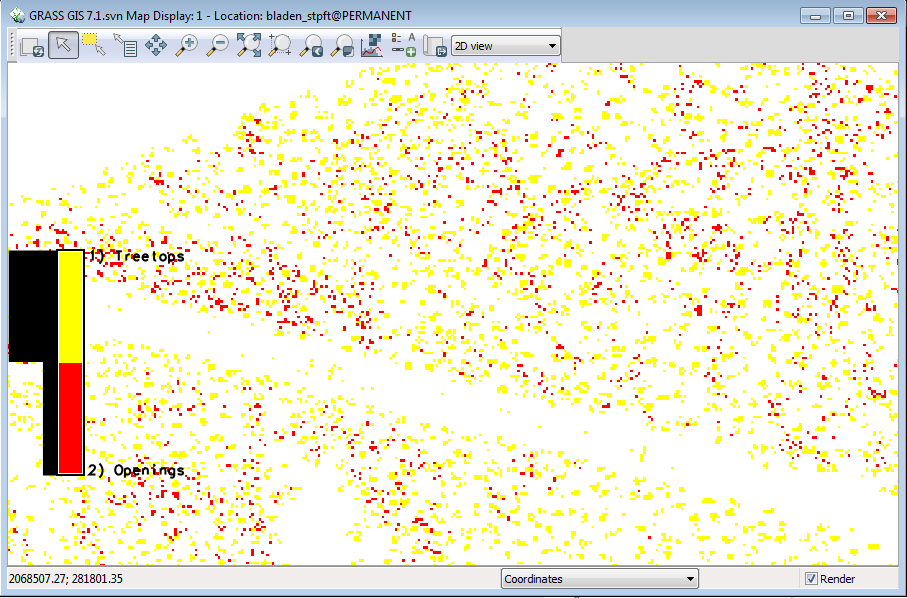
Your display should look like this ( zoomed into the carolina bay) | |||
[[File:Grasslidar_62.png| Treetops and Openings]]<br /> | |||
First, a note about r.reclass. It does not create a new layer, it just creates a virtual layer with a filter pointing to the orginal raster. This makes it very fast and saves on hard drive space. GRASS will treat it like a real layer. | |||
Second, notice the line of “canopy openings” where the transmission right of way going across the pocosin vegetation and the line in the upper left and the tree tops in the field beyond the ditch. We need to filter out by canopy height. | |||
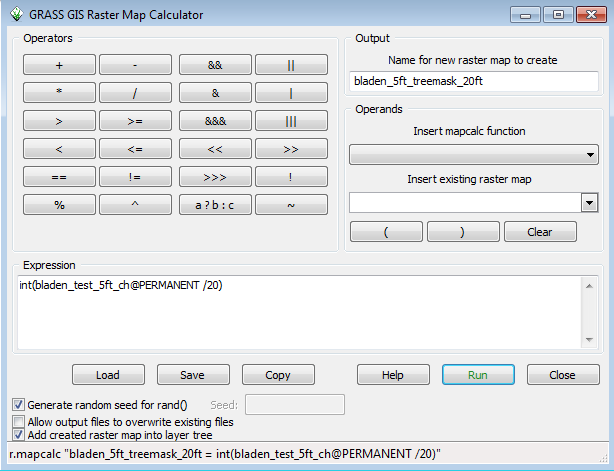
The best way to do that is to create a canopy height-based mask. In GRASS, the r.mask command, https://grass.osgeo.org/grass71/manuals/r.mask.html, is used. One issue is that our canopy height is a floating point raster and r.mask needs an integer raster. To fix this, we go back to r.mapcalc: | |||
* Start r.mapcalc | |||
* Select bladen_test_5ft_ch from the insert existing raster map layer dropdown | |||
* Click on the / button and type 20 | |||
* Type int( at the front of the equation and ) at the end | |||
Your mapcalc window will look like this: | |||
[[File:Grasslidar_63.png| Mapcalc treemask]]<br /> | |||
With | |||
int(bladen_test_5ft_ch@PERMANENT /20) | |||
as the equation in the Expression box | |||
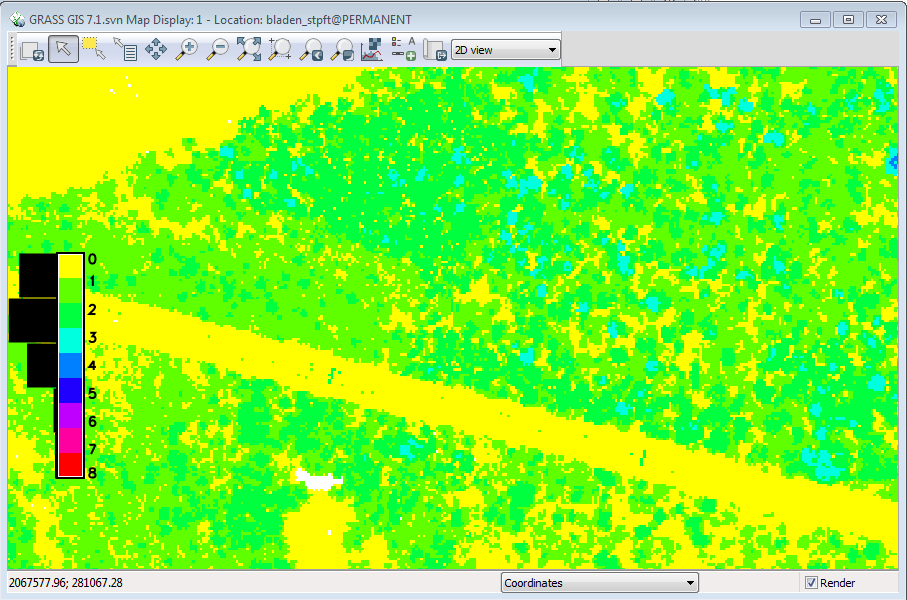
Your mask raster should look like this: | |||
[[File:Grasslidar_64.png| Height mask raster]]<br /> | |||
All we care about for tree tops are the areas in class 1 or higher, so start r.mask. | |||
* Select Raster-->Mask{r.mask] | |||
* In the Raster tab , Select bladen_5ft_treemask_20ft | |||
* In the box for Raster values to use for mask type: 1 thru 8 | |||
* Click Run | |||
[[File:Grasslidar_65.png| Mask Menu]]<br /> | |||
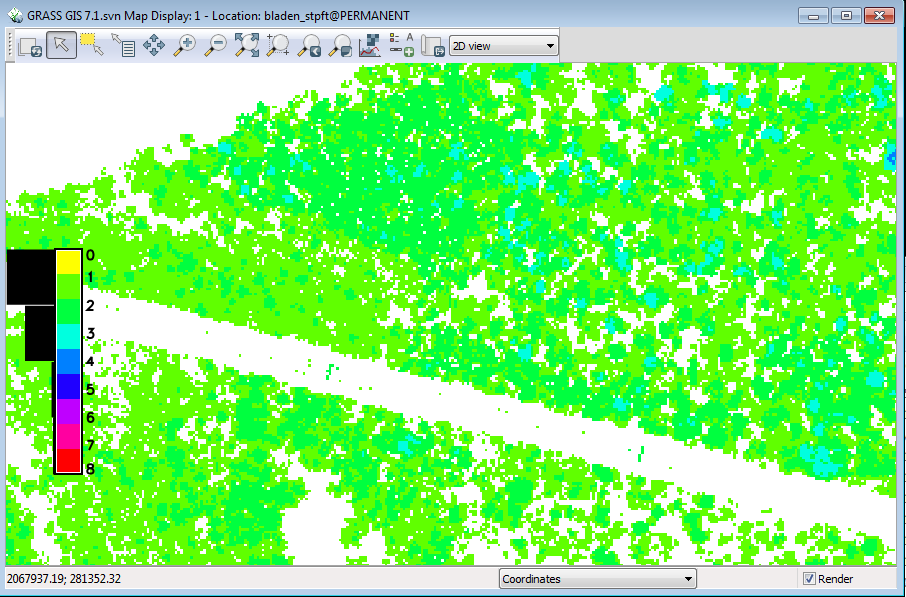
The same area now looks like this: | |||
[[File:Grasslidar_66.png| Mask Applied]]<br /> | |||
* Turn off the mask layer | |||
You should now see the treetops and canopy openings. Of the two classes here, the treetops are probably valid, but the canopy openings that show up with this mask are 20ft or more above the ground and are probably just low spots in the canopy rather than true canopy openings. | |||
[[File:Grasslidar_67.png| Mask Applied]]<br /> | |||
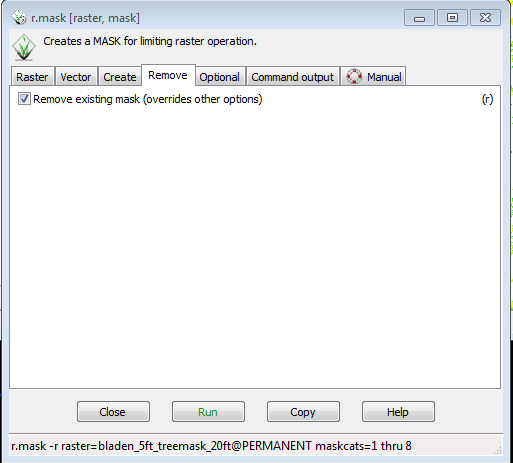
The raster mask will remain in place and affect all raster layers and raster operations until it is removed. To remove a raster mask: | |||
* Click on the Remove tab | |||
* Check the box for Remove existing mask | |||
* Click Run | |||
[[File:Grasslidar_68.png| Remove Mask ]]<br /> | |||
== Step 8: Exporting to GeoTiff == | |||
While GRASS is a rich environment for analysis, the GRASS native datasets are not easily accessible by other applications. Exporting to a common file format such as geotiff would make the data more generally available. | |||
To export the a raster layer from GRASS : | |||
* From the Layer Manager window select File-->Export Raster map-->Common export formats | |||
[[File:Grasslidar_69.png| Select Raster Export ]]<br /> | |||
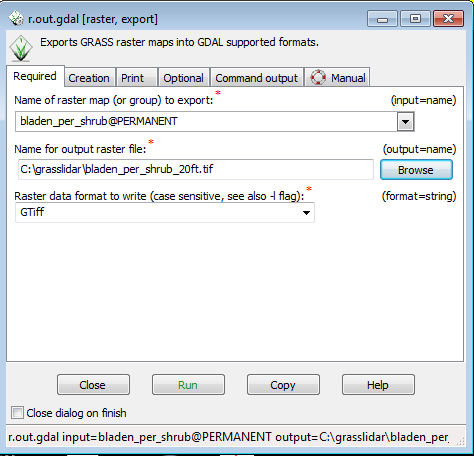
This will open the r.out.gdal menu, which uses the gdal library give you the option to write to many different raster formats. | |||
* Select bladen_per_shrub from the Name of raster map drop down | |||
* In the Name for output raster file option, Browse to the directory ( c:\grasslidar) and enter the name of the output file as bladen_per_shrub_20ft..tif | |||
* Keep the Raster data format as GTiff | |||
[[File:Grasslidar_70.png| R.out.gdal 1 ]]<br /> | |||
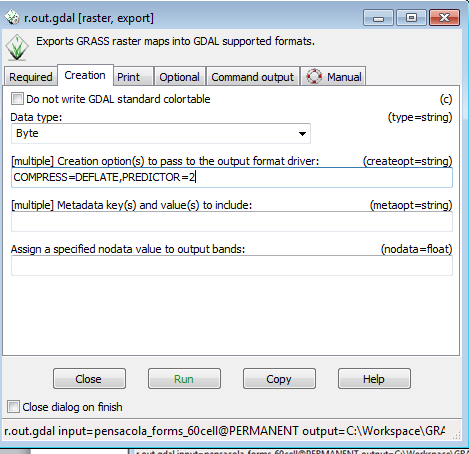
* Click on the Creation tab | |||
* Set the data type to byte ( 0-255 integer range) | |||
* In the Creation options window, type COMPRESS=DEFLATE,PREDICTOR=2 | |||
The compression options in the creation options will cause the output geotiff to be losslessly compressed using the deflate method. Predictor= 2 makes the compression more efficient. | |||
The export process finishes in about 2 seconds. | |||
[[File:Grasslidar_71.png| R.out.gdal 2 ]]<br /> | |||
== See also == | |||
* [[Processing lidar and UAV point clouds in GRASS GIS (workshop at FOSS4G Boston 2017)]] | |||
* [[Introduction to GRASS GIS with terrain analysis examples]] | |||
* [[From GRASS GIS novice to power user (workshop at FOSS4G Boston 2017)]] | |||
[[Category: Documentation]] | |||
[[Category: Tutorial]] | |||
Latest revision as of 21:51, 23 September 2018
This exercise was initially created as a session in a GIS training for the U. S. Fish and Wildlife Service in May, 2016 by Doug Newcomb.
Session Objectives
At the conclusion of this session, you will be able to:
- Open GRASS GIS and Create a Location from an existing file
- Link external raster elevation data to the Location
- Import LAS point cloud data to assess DEM accuracy
- Import LAS point cloud data to create various vegetation structure products
- Export raster vegetation structure data layers from GRASS to GeoTiff
Material Created By: Doug Newcomb (May 2016)
Software: GRASS 7.2
Directory Path: D:\grasslidar (assumed at some places, use any directory you want)
Image Files: D05_37_20026801_20141209.tif D05_37_20026803_20141209.tif D05_37_20026802_20141209.tif D05_37_20026804_20141209.tif
LAZ format LiDAR files: LA_37_20026801_20141209.laz LA_37_20026803_20141209.laz LA_37_20026802_20141209.laz LA_37_20026804_20141209.laz
Data can be accessed online here
Elevation data is commonly used in landscape analysis, but it is also quite useful in vegetation analysis. This exercise will walk you through basic analysis of a LiDAR point cloud to better understand vegetation structure.
All data are located in c:\grasslidar\data, unless otherwise noted.
Step 1: Creating GRASS Workspace
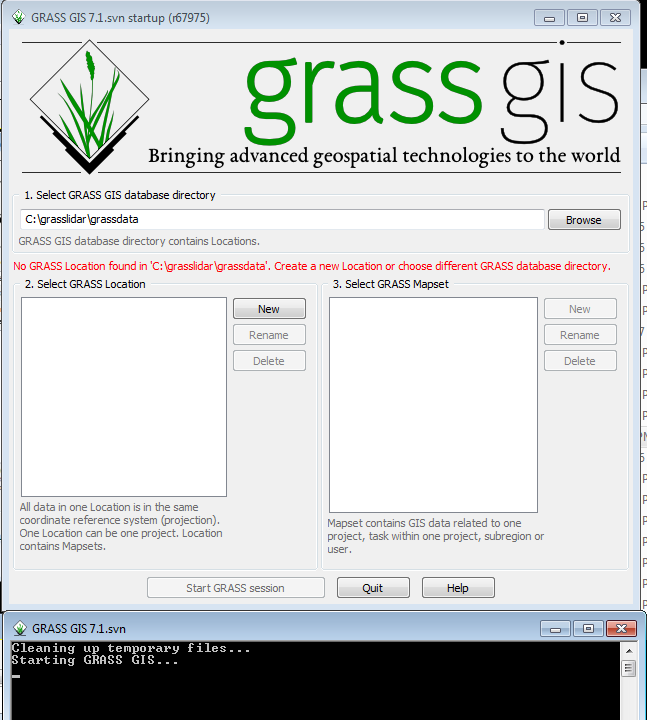
The first thing to do when starting to work in GRASS is to create a Location. GRASS Locations are single projection areas with a defined resolution and extent. The initial location can be easily created from an existing data set.
In Windows, Click on Start-->All Programs-->GRASS GIS 7.2--> GRASS GIS 7.2 GUI
Two windows will open, the GRASS startup window ( to select or create a workspace) and the GRASS command prompt.
- Click on the New button between the Location and Mapset windows. This will bring up the menu to define a new Location.
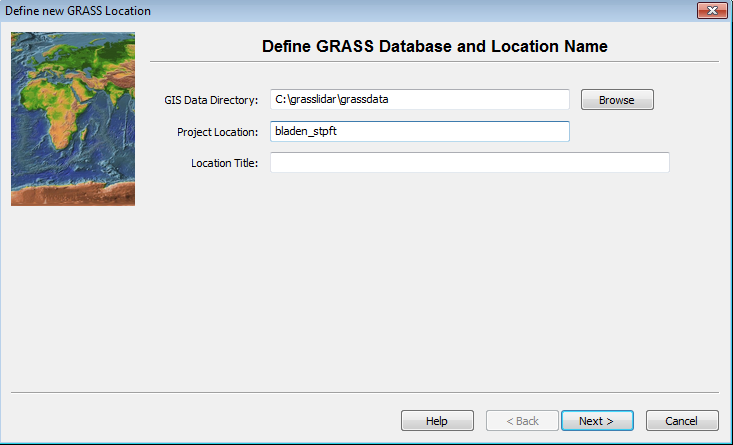
The GIS Data Directory is where all of your grass workspaces will reside. Creating a new directory with a unique name for grass data is recommended. This directory can be created anywhere that the user has write access. Project location is a subdirctory name for this particular project. Like with ArcGIS, it is best to avoid spaces in Directory names to avoid problems down the road. Click on the Browse button and select c:\grasslidar\grassdata. The data is from Bladen County, NC and is in the North Carolina State Plane NAD83(2011) projection with units feet, so call it bladen_stpft
- Enter the Data Directory and Project Location – this brings up the location creation method menu
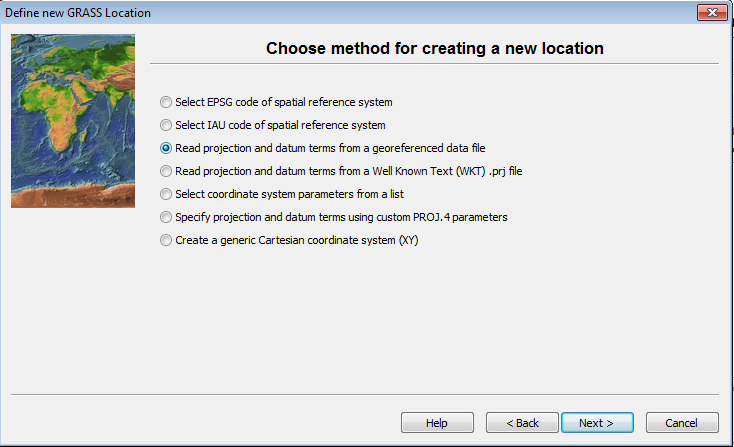
- Click on the radio button for Read projection and datum terms from a georeferenced file.
- Click Next
This brings up the georeferenced file dialog.
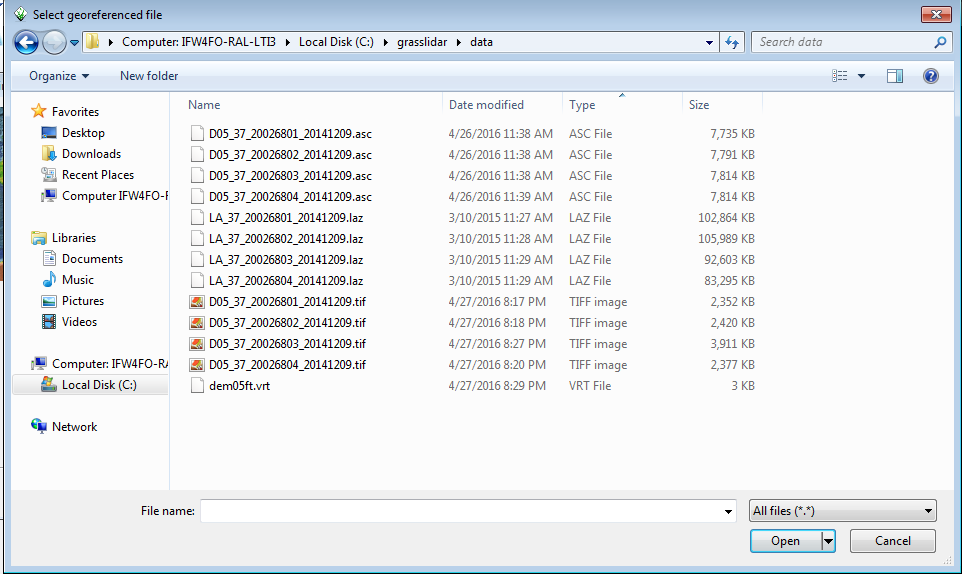
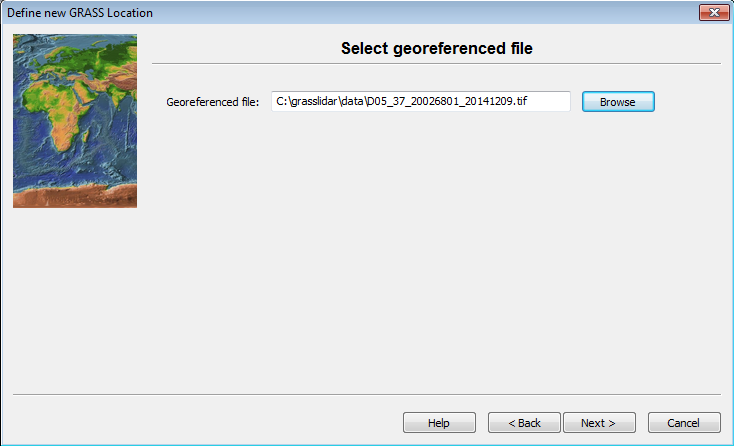
- Browse to C:\grasslidar\data and look at the data available:
In this directory, you will see 4 file types: asc, laz, tif, and vrt. LiDAR data is usually distributed as tiles of point clouds and Digital Elevation Models (DEMs) ASC and TIFF are common file formats to distribute DEM data. Asc is an ArcInfo ASCII Grid file, which is simply a text file representing a grid and does not have any projection data associated with it. Generally .tif files distributed as DEMs have projection information embedded in the file. LAZ is an open method of compression for LAS files. VRT is a virtual raster index of the 4 .tif files in this directory.
In this directory, and select D05_37_20026801_20141209.tif as the georeferenced file.
- Click Next.
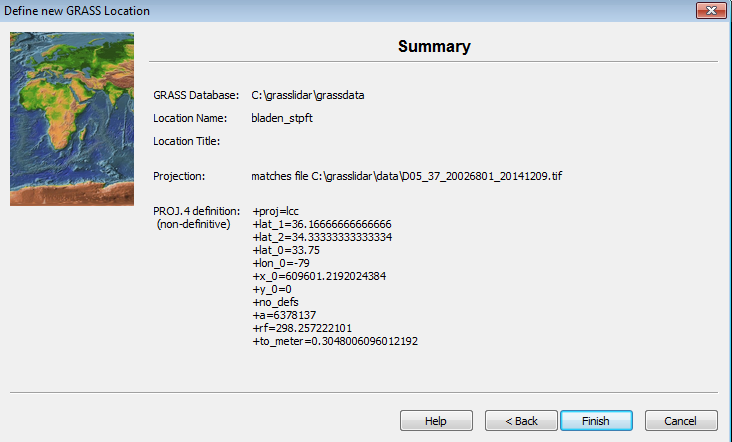
GRASS reads the georeference data from the elevation file and displays the projection data.
- Click Finish
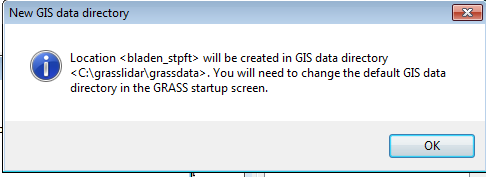
The next message relates to setting a default path for GRASS startup.
- Click OK
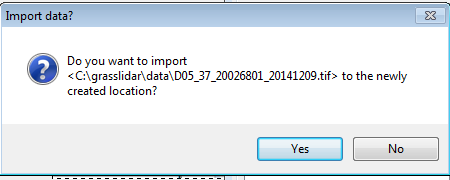
In the following message, you have the option if importing the data set you used for georeferencing the workspace.
- Click No
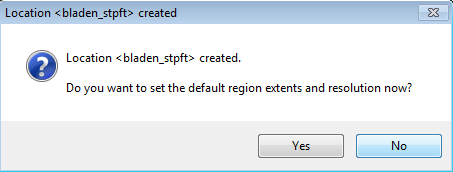
The location has been created. You are prompted to set the default region extents and resolution.
- Click No
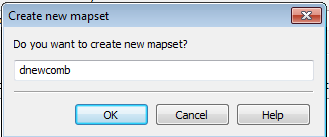
You are then prompted to create a new mapset.
- Click Cancel
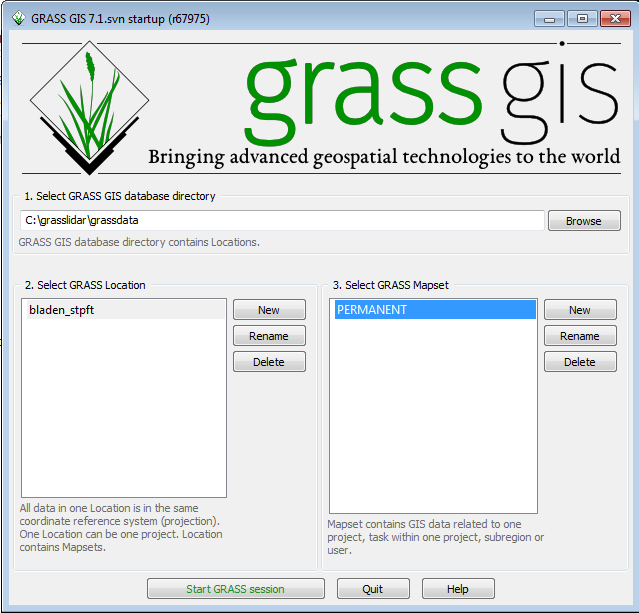
You have finished creating a Location with the default PERMANENT mapset. The next time GRASS is started, you can either select an existing Location and Mapset, create a new Mapset within the Location, or create a new Location.
- Click on the PERMANENT Mapset
- Click Start GRASS session
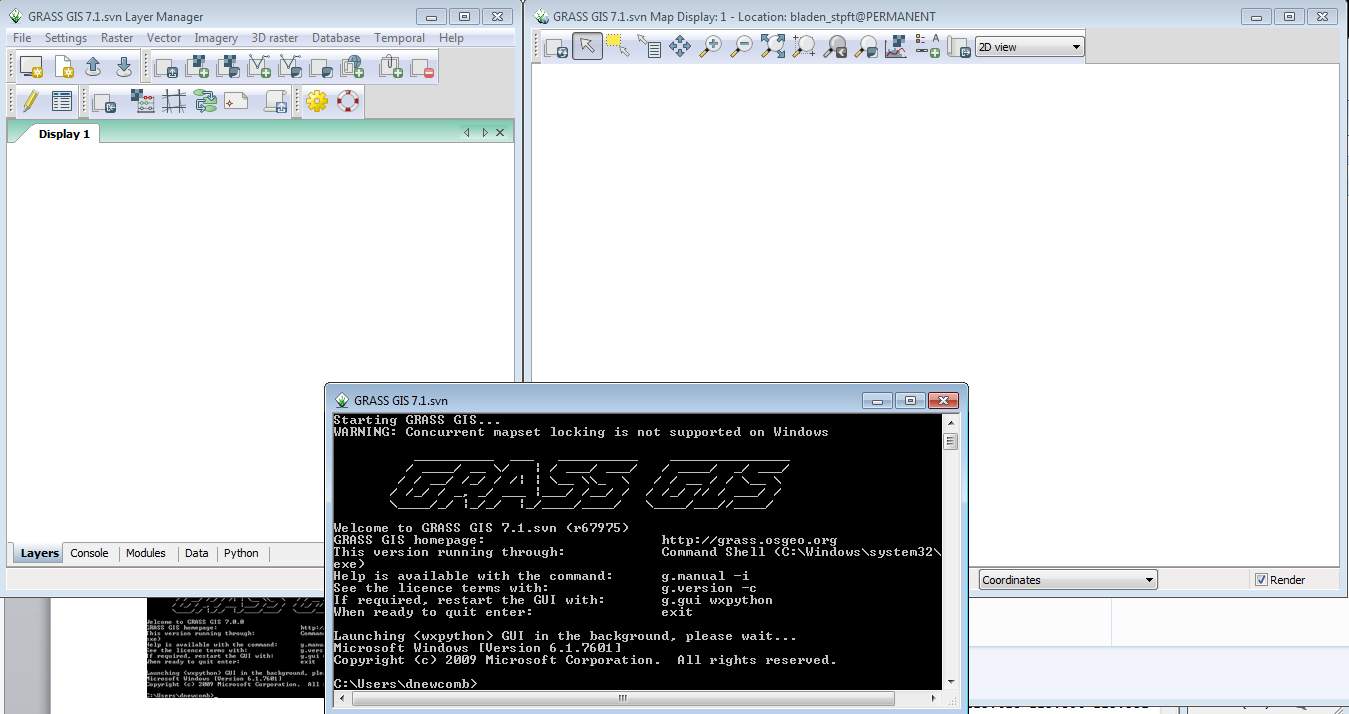
You will then see the Layer Manager Window, the Map Display Window for Display 1.
Step 2: Adding a data layer
We are now ready to add data to the Mapset. We can either import Raster and Vector data using the GDAL translation library, or we can link to existing external data sets read only to reduce data duplication. In this case, we will link to the 1m elevation data we used to create the Location using the r.external command.
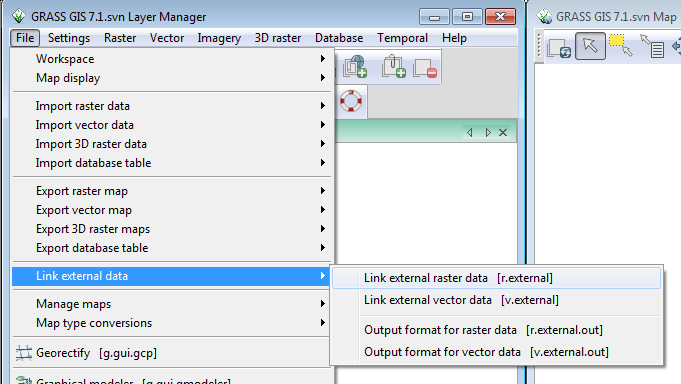
- In the Layer Manager window, click File-->Link external data-->Link external raster data.
This brings up the r.external dialog.
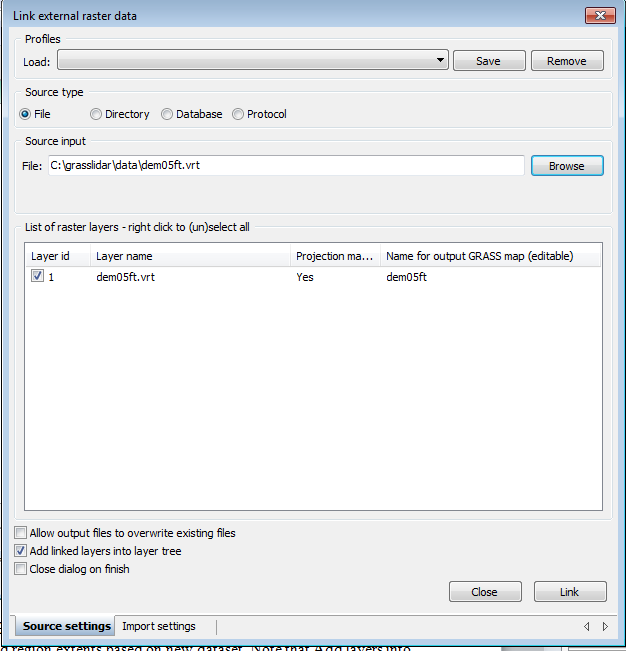
We would like to treat the 4 tiles of LiDAR and elevation data as a single seamless unit, to accomplish this, we are going to first select the demo05ft.vrt image index as our input raster.
- Browse to C:\grasslidar\data\dem05ft.vrt and select it. Note that the Add layers into layer tree is checked. For large data sets, leave this unchecked.
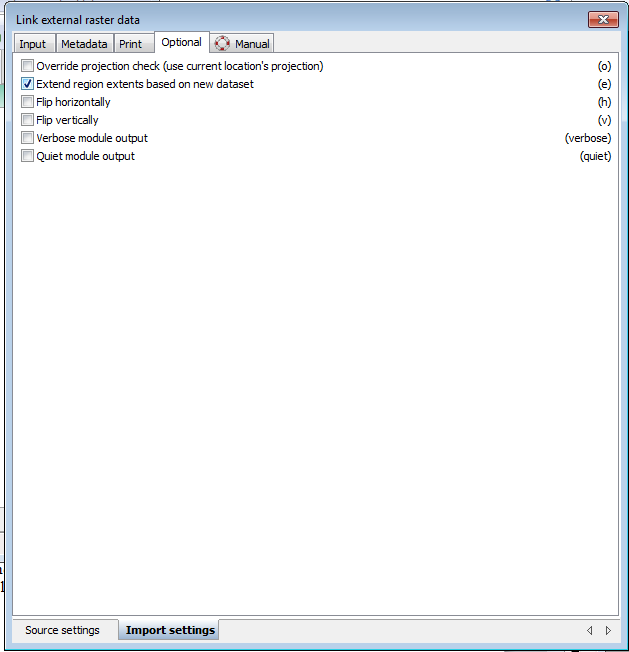
- Click on the import settings tab at the bottom
- Click on the box for Extend region extents based on new dataset.
- Click back to the Source settings tab and Click Link.
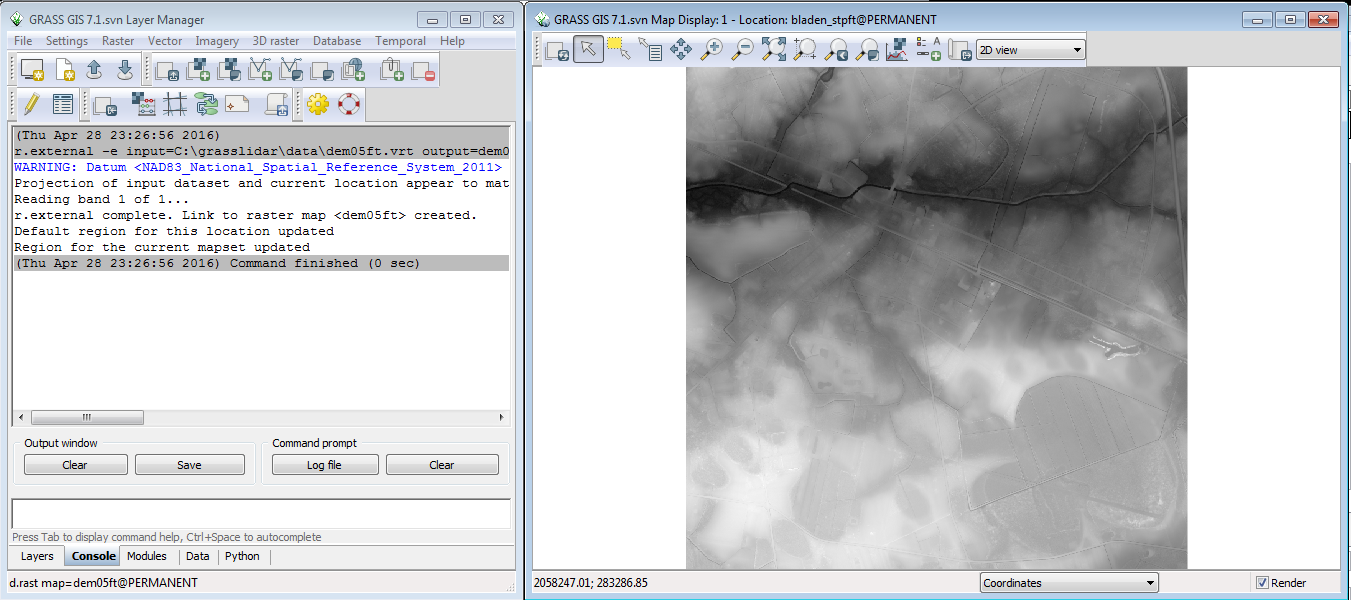
The Layer Manager window will switch to the Command console tab and display the results of the r.external command . Click the close button on the r.external dialog.
The 4 DEM rasters are displayed in the Map Display window as a single seamless data set . Any GIS software built with Geodata Abstraction Library(GDAL) ( including ArcGIS 10.0 and above) will see a vrt mosaic as a single raster layer. Creating the vrt is as simple as going to the black GRASS command window ,navigating to the directory in the with the rasters and typing: gdalbuildvrt dem05ft.vrt *.tif
For larger data sets, it takes much less time to link to a raster data set than import it and linking also saves on storage space.
Step 3: Setting up the region
The region in a GRASS mapset defines the extent and resolution of the results of any raster actions. To make sure that all of a raster layer is affected by a command, it is customary to set the region’s extent and resolution to match the raster layer. To set the region to match the elevation raster that we just linked to ,
- From the Layer Manager window drop down options, click the Settings-->Region-->Set region
- In the g.region menu, in the Set region to match raster map section, use the drop down to select dem05ft@Permanent.
- Click Run
When the command finishes, your will see “Command finished” and the time taken in the g.region window .
- Click Close in the g.region window
- Click Settings-->Region-->Display region
In the command console tab of the Layer Manager window, you see the current region definintion: resolution, bounds, rows and columns. The output resolution of raster layers created will usually match the region resolution. To change the output resolution, you will need to change the resolution of the region.
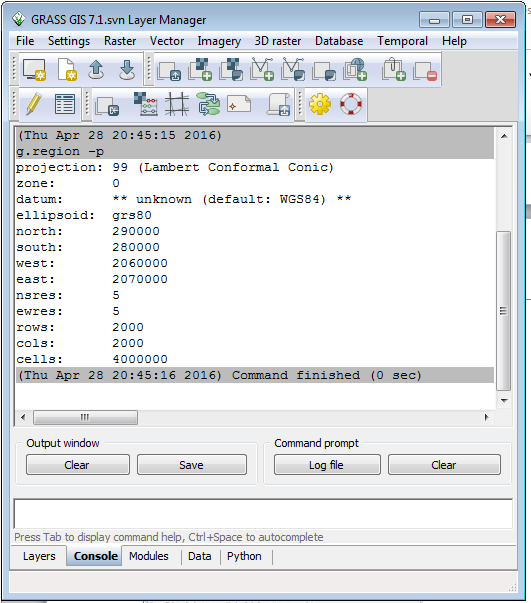
A quick note about the results. The datum is not recognised and assumed to be WGS84 by GRASS. The datum is NAD83 (2011). The data coordinates have not changed, the datum information has not been recognized on import. We can work with the data and assign the correct datum information after export from GRASS. This should be corrected by the final release of this version of GRASS.
Step 4: Processing the Lidar data
There are 4 lidar tiles that correspond to the 4 tiles of elevation data. The data is in the Compressed LAS files (laz) format. LASZip, http://www.laszip.org/, is an open compression method that losslessly compresses LAS format LiDAR data files to about 25 -30 percent of the original size.
You will be using the GRASS 7.1 r.in lidar. R.in.lidar uses a binning process, performing an analysis on the points that fall in each raster cell. It’s best to process las tiles as a single unit to avoid border effects. There are two ways to do this: 1) Merge the las files together into a single file using tools such as lastools lasmerge, https://rapidlasso.com/lastools/lasmerge/ . Many software packages are still limited to LAS format 1.2 which can only handle up to 4.2 billion points per file ( a current limitation in GRASS7.1) . This option can also take up hard drive space. 2) The second option available in r.in.lidar is to give a filename with a list of adjacent LAS/LAZ files and it will treat it as a virtual seamless LiDAR data set. If you look in the data directory, you will see laslist.txt. The contents of the file are: C:\grasslidar\data\LA_37_20026801_20141209.laz C:\grasslidar\data\LA_37_20026802_20141209.laz C:\grasslidar\data\LA_37_20026803_20141209.laz C:\grasslidar\data\LA_37_20026804_20141209.laz
This is a list of the compressed las files in the directory that you will be processing. Give r.in.lidar c:\grasslidar\data\laslist.txt as the input file list, and it will treat all 4 files as a single point cloud. This method has been tested on file lists up to 966 las files.
This exercise will use the second method. To start r.in.lidar, go to File-->Import raster data-->LAS Lidar points import.
This will bring up the r.in.lidar menu.
You will notice that there are several tabs across the top of the command window. The first tab is the Input tab . You can either enter the name of the individual las or laz file, or you can enter the name of a txt file with a list of las or laz files. In the second option, browse to the location of the laslist.txt file and select it as shown.
Next we will set the output raster layer.
- Click on the Output tab
- In the Name for output raster layer box, type “ bladen_test_5ft_pt_count” as the output layer
You will be binning the data using a 5ft resolution cell. In this case, you will be counting how many LiDAR returns are in each 5ft cell.
- Click on the Statistic tab
- Select n from the Statistic to use for raster values drop down menu
- Click on the selection tab
- You want all points except the low noise ( class 7) so put 1,2,3,4,5,6,8,9,10,11,12,13,14,15,16,17,18,19,20 into the box for “Only import points of selected classes.” (feel free to copy and paste)
- Click on the Optional tab
- Select the check box for Override projection check
- Click Run.
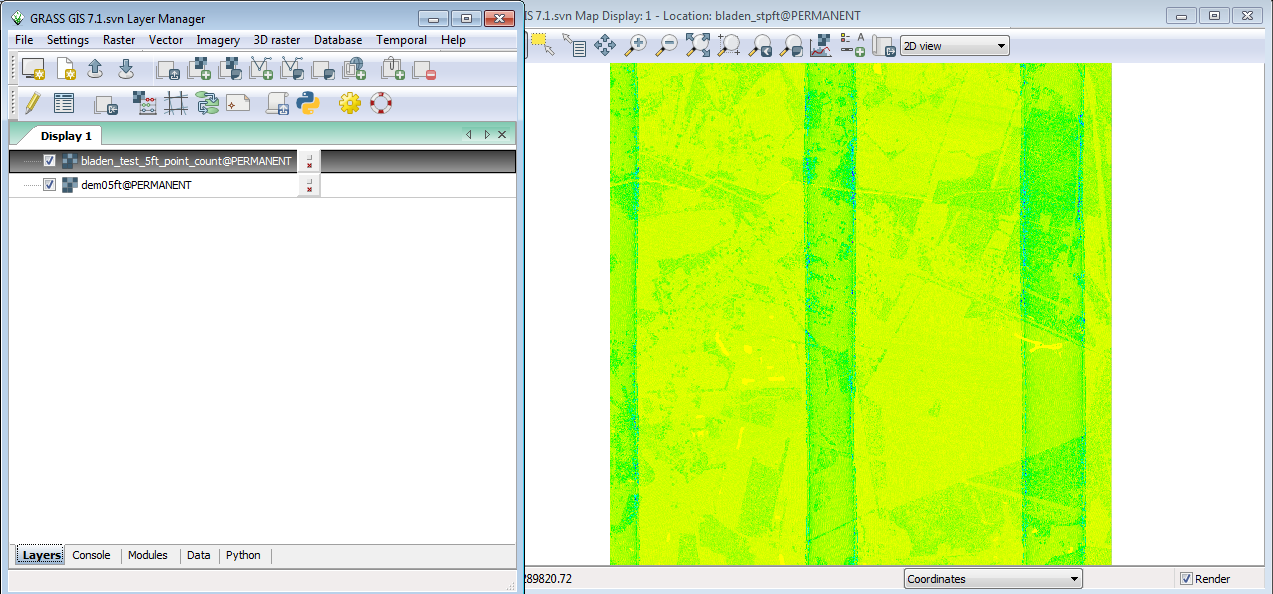
Your display should look like this, with the result of the point count automatically added to the display.
To add a legend to the display:
- Click on the Add map elements button on the top of the Display window
- Select show/hide legend
The default legend is not clear for this data set, so simplify the legend by:
- Double clicking on the legend to bring up the d.legend menu.
- Click on the Gradient tab and check the Draw smooth gradient and Add histogram to smoothed legend.
- Change the number of text labels from the default of 5 to 8.
- Click OK
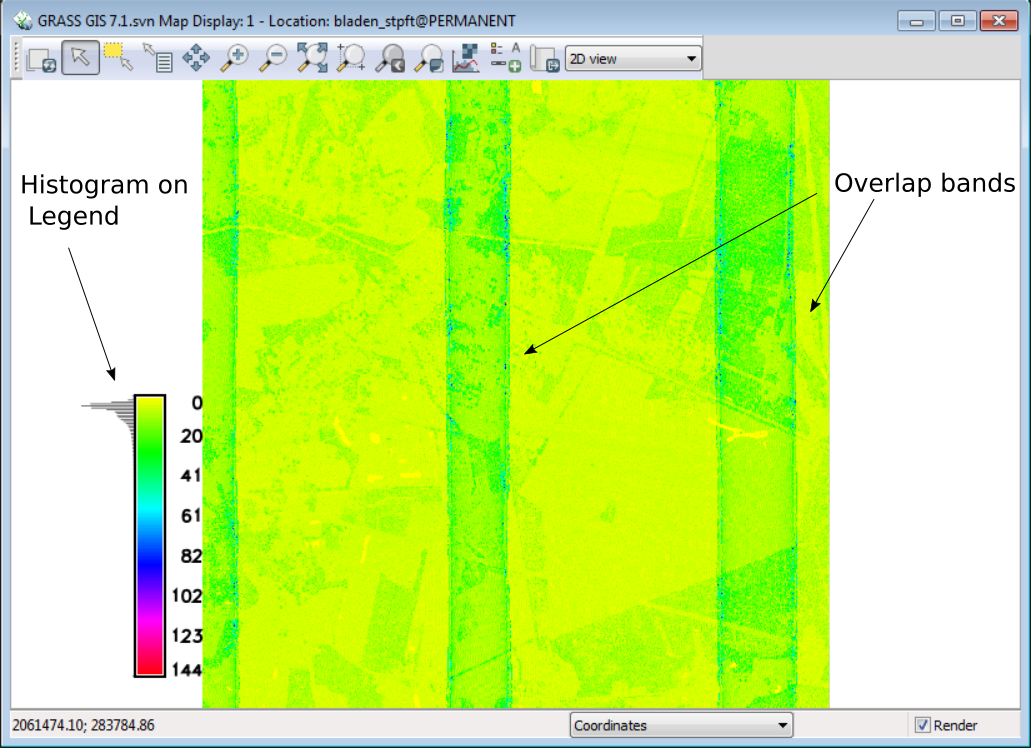
As you can see in the illustration above, the histogram on the legend tells you that most of the cells have fewer than 10 points per cell. For any valid statistical analysis of the point values in the cell you would need to pick a cell size larger than 5ft if you are performing statistics on the point values within each cell.
You can also see the overlap of the LiDAR data collection flight lines. This makes the use of direct point count density useless as a measure of vegetation density unless all overlap points are removed.
Step 6: Canopy Heights and Other Forest Structure Metrics
Calculation of canopy heights involves determining the distance between the ground and the top of the vegetation. It can be done two steps , creating a digital surface model (DSM) and subtracting the Digital Elevation Model (DEM) With GRASS, this can be done in one step.
Go back to the r.in.lidar menu:
- Set the output raster to bladen_test_5ft_ch.
- Go to the statistic tab and select max from the drop down menu.
- Select the Transform tab
- From the Subtract raster values from the z coordinates, select dem05ft from the dropdown menu.
- Go to the Optional tab
- Check the boxes next to Override projection check and Use base raster actual resolution instead of computational region
- Click Run
- Select the Display window and double-click on the legend.
- Select as the input raster layer bladen_test_05ft_ch
You have just used the 5 ft resolution DEM to convert the z values of the point cloud to height above ground. Selecting the maximum value in each cell now gives the canopy height in each cell. Using this proceedure allows you to convert the LiDAR point Z values to height above ground at the 5ft resolution, but the calculations after the conversion will be done for the cell size of the current resolution of the location.
Look back at the legend. You will notice that the bottom value is negative 4. Why is that? Run r.in.lidar again with the statistic st to min and the output raster set to bladen_test_05ft_min.
- Go back to d.legend and set the raster to bladen_test_05ft_min.
- Click on the Gradient tab and set the number of text labels to 20
- Click apply
You can see that the bulk of the minimum values are less than the DEM surface. Turn off all layers except the DEM layer. Zoom into the large oval area in the Southeast corner of the DEM.
This feature is called a Carolina bay. The northern half has been ditched and drained, and the southern half remains in the native thick vegetation. Turn the min values back on.
- Go to raster-->Manage colors --> Color tables. This will open the r.colors menu.
- Select the Define tab and in the Name of color table dropdown select elevation.
You will notice that in the light green and yellowish areas are significantly above the DEM surface. This is an indication that the laser did not penetrate the vegetation to hit the ground. While the DEM is not perfect, it gives us a better ground surface than just using the min value. There are ways to work around those differences, but that is not the topic for today.
Look at the other end
- Set the region to match the 5ft dem
- Remove the base raster in the Transform tab and in the Optional tab
- Remove the zrange on the Selection tab
- Set the output raster to top_of_canopy_5ft
- Run r.in.lidar with the max method
Change your legend to match the top of canopy layer. What is the max height that you see?
- Click Raster-->Reports and statistics-->Basic raster metadata (r.info)
- Select dem05ft
- Click Run
R.info gives basic information about the raster layer. You see the rows, columns, number of cells, projection, the command that created the raster is listed as well. This is all useful information for metadata construction. The min and max values of the raster is what you need to look at now. The maximum value in the DEM is 2264.559. Since we have no trees in NC over 200ft, it seems we have some invalid high points. To remedy this:*
- Go back to the Selection tab and and
- for the zrange enter -10,250.
- Click Run
This gives a -10, 250 range around the high and low ground in the area (93-10 =83 and 128+250 = 378). This should exclude most of the extraneous points and leave a 5 ft resolution Digital Surface Model(DSM). Change the legend to display the new raster layer. What differences do you see?
For now we are going to leave this dataset, but we will come back to it later.
- Go back to Settings-->Region--> Set region to open the g.region dialog.
- Select the Resolution tab.
- Enter 20 in the 2D grid resolution box
- Click Run
If you go to Settings-->Region-->Display region You will see : nsres: 20 ewres: 20
The Region resolution setting is now for 20ft cells.
- Go back to the r.in.lidar menu and:
- Set the output raster to bladen_test_20ft_ch and
- Set the statistic to max.
- Set the transform layer back to dem05ft
- Check the box for use base raster resolution
- Click Run
- Set the legend to bladen_test_20ft_ch
The Z values were converted to height above ground using the 5ft DEM and then statistics were calculated for 20ft cells. Notice how you see the outlines of the ditches.
- Make sure that the bladen_test_20ft_ch is highlighted
- Click on the Query selected raster/vector button ( 4th from the left in the display window)
- Place the crosshairs on one of the ditches
- Left Click
A window will pop up giving the location coordinates, the layer being queried, the color of the feature, and the value of the feature. In this case, the canopy height of the feature. The Z conversion to height above ground is not perfect and may depend on the accuracy of the DEM and the steepness of the slope represented by the raster.
Remember that a slope is a smooth surface that is being interpolated to a grid format.
Choosing a cell size can be important. At a 20ft resolution, you will be getting parts of larger trees and 60ft may be more appropriate to capture a more completepicture of the landscape vegetaion. Converting to height above ground allows for the use of cells of arbitrary size. Your analysis can match up with the cell size and footprint to exactly match the cell alignment of existing data.
There are several other statistical measures that are available in r.in lidar: n, mean, range, sum, standard deviation, variance, coefficeint of variance, median,percentile, skewness, trimmean. Mean height, variance and skewness are metrics used in vegetation analysis.
At the 20ft resolution, repeat r.in.lidar for variance and mean methods. What differences do you see in the resulting rasters?
Repeat r.in.lidar at 20ft resolution using n as the statistic to create a point count layer.
- Go to the Selection tab and
- Set the z range to 3,10
- Set the output raster to bladen_20ft_shrub_count
- Click Run
- Go to the Selection tab
- Set the zrange to 10,20
- Set the output raster to bladen_20ft_midstory_count
- Click Run
As stated above, using the raw point count is not suitable for vegetation analysis in areas where there are overlaps in collection. However, if horizontal sections of the point counts per cell are divided by the total point count per cell you can compare the percentage of the points that occur in horizontal sections. In this case, we will be using raster algebra to divide the shrub and midstory point counts by the total point counts using r.mapcalc, https://grass.osgeo.org/grass71/manuals/r.mapcalc.html .
- Go to Raster-->Raster map calculator (r.mapcalc)
This will open the r.mapcalc menu:
- Enter bladen_per_shrub in the Name for new raster map to create
- Use the dropdown in the Insert existing raster map section to select bladen_20ft_shrub_count
- Click on the / button for division
- Use the dropdown to select bladen_20ft_count
Now things get interesting. The point count rasters are integers. Mathmatical operations between integers results in an integer. Dividing an integer by a larger integer number results in a number less than one. In an integer grid, this truncates to 0.
To get around this, you convert each integer grid before the operation to a floating point grid, so bladen_20ft_shrub becomes float(bladen_20ft_shrub) and bladen_20ft_count becomes float(bladen_20ft_count) and you divide one floating point grid by another floating point grid and you get a floating point grid.
Multiply this floating point grid by 100 to put the values back in the range of 100 to 0. Finally, convert results of the entire operation back to an integer grid by enclosing it with int() . The final command in the Expression box is:
int((float(bladen_20ft_shrub_count@PERMANENT)/float(bladen_20ft_count@PERMANENT))*100)
The resulting raster should look like this:
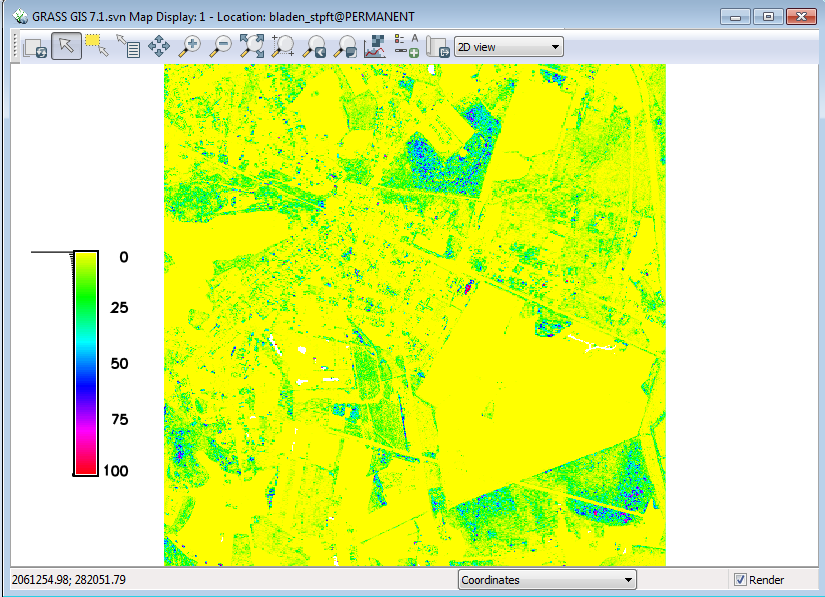
Repeat the exercise with the midstory layer and compare the two layers. Do you see differences between the shrub and midstory percentage of points layers?
We will continue the vegetation analysis, but first we need to take a short side trip into terrain analysis that will relate back to our vegetation analysis.. Terrain analysis will be done using geomrphons.
Step 5: Terrain analysis with geomorphons
What are geomorphons? Geomorphon is a new technique for classifying terrain based on a line of sight analysis, see http://geomorphometry.org/system/files/StepinskiJasiewicz2011geomorphometry.pdf , and http://www.sciencedirect.com/science/article/pii/S0169555X12005028
The technique uses a line of sight metric for terrain form calculation. There are options for a simple 10 class representation, a 498 class numeric coding, or positive and negative encoding per cell on line of sight.
For this activity, we will use the simple 10 class calculation. Outputs will be cells with the following classes: Flat, peak, ridge, shoulder, spur, slope hollow, footslope, valley, and pit.
To do our terrain classification, we will need to load the r.geomorphon addon to GRASS.
GRASS has an online repository of Addons much like QGIS has for plugins. Most addons do not require administrative access to install.
- In the Layer Manager window, click on Settings-->Addons extentions-->Install extention from addons
This will open the Fetch and install extension window. GRASS commands are named by their function: r.* commands are raster commands, v.* commands are vector commands, i.* commands are image analysis commands.
- Click on the + next to raster to expand the view on all raster commands
- Select r.geomorphon
- Click install
In the Layer Manager window you can follow the progress of the installation.
In this case, downloading and installing the addon took 4 seconds on a 2010 vintage Windows 7 laptop.
Step 6: Running r.geomorphon
The r.geomorphon command documentation is at http://grass.osgeo.org/grass70/manuals/addons/r.geomorphon.html and the publications for the technique can be found at the end of the command description.
To start r.geomorphon,
In the GRASS Command window, type r.geomorphon and press Enter.
Entering the command without arguments brings up the r.geomorphon menu. You will notice that there are tabs across the top. The first tab is the Required Tab, with required data inputs to run the command.
- From the dropdown for Input DEM, dem05ft@PERMANENT ( remember, the current resolution for the region is 20ft. GRASS will take the center value of the 5ft DEM in each 20ft cell and use that for the 20ft cell value, your output raster will be 20ft resolution)
- In the Outer search radius, enter 30 ( number of cells around each cell to analyze) This is a very flat area, so the number may need to be higher to extract some features.
You will note that as the command arguments are entered, they are echoed across the bottom.
- Click on the patterns tab.
- Type common_20ft_30cell in the box under Most common geomorphic forms.
- Click Run
The tab will immediately switch to the Command Output tab, the command options will be echoed , and a progress bar will show the progress of the command. You will need to manually add the result to the Display.
- At the top of the Layer Manager window, Click Add raster map layer button
- In the d.rast menu that pops up, select common_20ft_30cell
- Click OK
Your results should look like this ( once you select the legend to show the current raster):
A check of the Windows Task Manager shows that the r.geomorphon command is using less than 100 MB of RAM to process the area. This process will take 7.5 minutes on a 2010 vintage i7 laptop.
Larger areas and larger cell radius extents will require more memory. This process scales fairly well. It has been used on the Statewide 20ft elevation grid for North Carolina ( 6.6 billion cells) with a radius of 30 cells, but it required more than 4GB of RAM and took overnight to run on a Linux workstation. GRASS 7.2 on Windows is 64 bit and will address more than 4 GB of RAM on that platform.
You can see a lot of different patterns, but what do they mean? Click on the Query raster / vector maps tool and you can click individual cells to extract the value.
For example, a blue pixel represents integer value 9 or valley.
Alternatively, you can
- Click on the Add map elements Button
- Select Show/Hide legend
- Click on the Gradient tab
- Check the boxes for draw smooth Gradient and ad histogram
- Put 10 in Number of text labels
- Click Apply
You should then see the following legend with the integer values, classes and histogram by class.
Pretty handy way of describing the landscape. What does this have to do with vegetation?
Step 7: Extracting canopy features using geomorphons
- Add the bladen_test_5ft_ch ( add raster) .
- Set the region to match bladen_test_5ft_ch(SettingsRegionSet regionSet region to match raster map)
- Run a 3 cell common geomorphon analysis on the 5ft canopy height
We want to look at the top of the canopy to pick out treetops and canopy openings. The treetops should be summits and the canopy openings should be depressions. We don’t care about any other class, so let’s create a new layer with just those two classes.
- Go to Raster-->Change category values and classes-->Reclassify (r.reclass)
- Select common_5ft_ch_3cell for the Name of raster to be reclassified.
- Set the output raster to treetops_openings_5ft
- Go down to the enter values directly box and type in:
1 = Null 2 = 1 Treetops 3 thru 9 = NULL 10 = 2 Openings
- Turn off all other layers
- Set your legend to treetops_openings_5ft
Your display should look like this ( zoomed into the carolina bay)
First, a note about r.reclass. It does not create a new layer, it just creates a virtual layer with a filter pointing to the orginal raster. This makes it very fast and saves on hard drive space. GRASS will treat it like a real layer.
Second, notice the line of “canopy openings” where the transmission right of way going across the pocosin vegetation and the line in the upper left and the tree tops in the field beyond the ditch. We need to filter out by canopy height.
The best way to do that is to create a canopy height-based mask. In GRASS, the r.mask command, https://grass.osgeo.org/grass71/manuals/r.mask.html, is used. One issue is that our canopy height is a floating point raster and r.mask needs an integer raster. To fix this, we go back to r.mapcalc:
- Start r.mapcalc
- Select bladen_test_5ft_ch from the insert existing raster map layer dropdown
- Click on the / button and type 20
- Type int( at the front of the equation and ) at the end
Your mapcalc window will look like this:
With int(bladen_test_5ft_ch@PERMANENT /20) as the equation in the Expression box Your mask raster should look like this:
All we care about for tree tops are the areas in class 1 or higher, so start r.mask.
- Select Raster-->Mask{r.mask]
- In the Raster tab , Select bladen_5ft_treemask_20ft
- In the box for Raster values to use for mask type: 1 thru 8
- Click Run
The same area now looks like this:
- Turn off the mask layer
You should now see the treetops and canopy openings. Of the two classes here, the treetops are probably valid, but the canopy openings that show up with this mask are 20ft or more above the ground and are probably just low spots in the canopy rather than true canopy openings.
The raster mask will remain in place and affect all raster layers and raster operations until it is removed. To remove a raster mask:
- Click on the Remove tab
- Check the box for Remove existing mask
- Click Run
Step 8: Exporting to GeoTiff
While GRASS is a rich environment for analysis, the GRASS native datasets are not easily accessible by other applications. Exporting to a common file format such as geotiff would make the data more generally available.
To export the a raster layer from GRASS :
- From the Layer Manager window select File-->Export Raster map-->Common export formats
This will open the r.out.gdal menu, which uses the gdal library give you the option to write to many different raster formats.
- Select bladen_per_shrub from the Name of raster map drop down
- In the Name for output raster file option, Browse to the directory ( c:\grasslidar) and enter the name of the output file as bladen_per_shrub_20ft..tif
- Keep the Raster data format as GTiff
- Click on the Creation tab
- Set the data type to byte ( 0-255 integer range)
- In the Creation options window, type COMPRESS=DEFLATE,PREDICTOR=2
The compression options in the creation options will cause the output geotiff to be losslessly compressed using the deflate method. Predictor= 2 makes the compression more efficient. The export process finishes in about 2 seconds.