R statistics/spgrass6: Difference between revisions
(Moved here from R_statistics) |
mNo edit summary |
||
| Line 1: | Line 1: | ||
'''This page refers to the usage of R within a GRASS GIS 6 session.''' | '''This page refers to the usage of R within a GRASS GIS 6 session.''' (see also [[R_statistics/rgrass7]]) | ||
=== Startup === | === Startup === | ||
Revision as of 14:29, 19 July 2015
This page refers to the usage of R within a GRASS GIS 6 session. (see also R_statistics/rgrass7)
Startup
- First start a GRASS session. Then, at the GRASS command line start R.
- In this example we will use the sample Spearfish dataset.
Reset the region settings to the defaults
GRASS> g.region -d
Launch R from the GRASS prompt
GRASS> R
Load the spgrass6 library:
> library(spgrass6)
Get the GRASS environment (mapset, region, map projection, etc.); you can display the metadata for your location by printing G:
> G <- gmeta6()
Listing of existing maps
List available vector maps:
execGRASS("g.list", parameters = list(type = "vect"))
List selected vector maps (wildcard):
execGRASS("g.list", parameters = list(type = "vect", pattern = "precip*"))
Save selected vector maps into R vector:
my_vmaps <- execGRASS("g.list", parameters = list(type = "vect", pattern = "precip*"))
attributes(my_vmaps)
attributes(my_vmaps)$resOut
List available raster maps:
execGRASS("g.list", parameters = list(type = "rast"))
List selected raster maps (wildcard):
execGRASS("g.list", parameters = list(type = "rast", pattern = "lsat7_2000*"))
Reading in data
Read in two raster maps (Spearfish sample dataset):
> spear <- readRAST6(c("geology", "elevation.dem"),
cat=c(TRUE, FALSE), ignore.stderr=TRUE,
plugin=NULL)
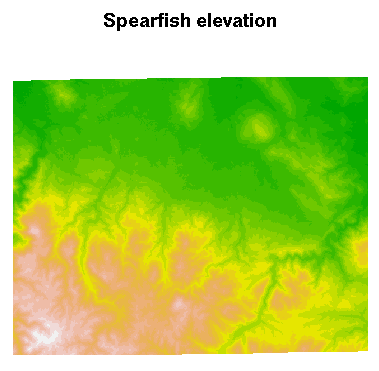
The metadata are accessed and available, but are not (yet) used to structure the sp class objects, here a SpatialGridDataFrame object filled with data from two Spearfish layers. Here is a plot of the elevation data:
> image(spear, attr = 2, col = terrain.colors(20))
Add a title to the plot:
> title("Spearfish elevation")
In addition, we can show what is going on inside the objects read into R:
> str(G)
List of 26 $ GISDBASE : chr "/home/rsb/topics/grassdata" $ LOCATION_NAME: chr "spearfish57" $ MAPSET : chr "rsb" $ DEBUG : chr "0" $ GRASS_GUI : chr "text" $ projection : chr "1 (UTM)" $ zone : chr "13" $ datum : chr "nad27" $ ellipsoid : chr "clark66" $ north : num 4928010 $ south : num 4913700 $ west : num 589980 $ east : num 609000 $ top : num 1 $ bottom : num 0 $ nsres : num 30 $ nsres3 : num 30 $ ewres : num 30 $ ewres3 : num 30 $ tbres : num 1 $ rows : int 477 $ rows3 : int 477 $ cols : int 634 $ cols3 : int 634 $ depths : int 1 $ proj4 : chr "+proj=utm +zone=13 +a=6378206.4 +rf=294.9786982 +no_defs +nadgrids=/home/rsb/topics/grass61/grass-6.1.cvs/etc/nad/conus"
> summary(spear)
Object of class SpatialGridDataFrame
Coordinates:
min max
coords.x1 589980 609000
coords.x2 4913700 4928010
Is projected: TRUE
proj4string : [+proj=utm +zone=13 +a=6378206.4 +rf=294.9786982 +no_defs +nadgrids=/home/rsb/topics/grass61/grass-6.1.cvs/etc/nad/conus]
Number of points: 2
Grid attributes:
cellcentre.offset cellsize cells.dim
1 589995 30 634
2 4913715 30 477
Data attributes:
geology elevation.dem
sandstone:74959 Min. : 1066
limestone:61355 1st Qu.: 1200
shale :46423 Median : 1316
sand :36561 Mean : 1354
igneous :36534 3rd Qu.: 1488
(Other) :37636 Max. : 1840
NA's : 8950 NA's :10101
Summarizing data
We can create a table of cell counts:
> table(spear$geology)
| metamorphic | transition | igneous | sandstone | limestone | shale | sandy shale | claysand | sand |
|---|---|---|---|---|---|---|---|---|
| 11693 | 142 | 36534 | 74959 | 61355 | 46423 | 11266 | 14535 | 36561 |
And compare with the equivalent GRASS module:
> execGRASS("r.stats", flags=c("c", "l"), parameters=list(input="geology"), ignore.stderr=TRUE)
1 metamorphic 11693 2 transition 142 3 igneous 36534 4 sandstone 74959 5 limestone 61355 6 shale 46423 7 sandy shale 11266 8 claysand 14535 9 sand 36561 * no data 8950
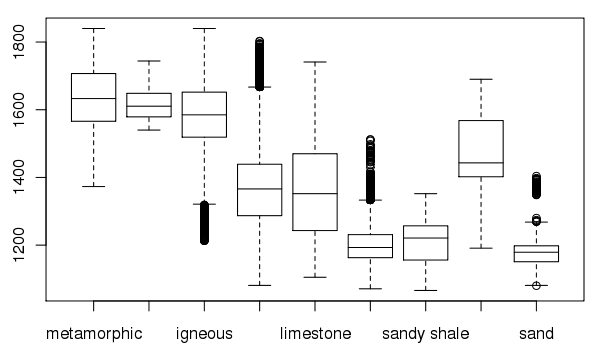
Create a box plot of geologic types at different elevations:
> boxplot(spear$elevation.dem ~ spear$geology, medlwd = 1)
Exporting data back to GRASS
Finally, a SpatialGridDataFrame object is written back to a GRASS raster map:
First prepare some data: (square root of elevation)
> spear$sqdem <- sqrt(spear$elevation.dem)
Export data from R back into a GRASS raster map:
> writeRAST6(spear, "sqdemSP", zcol="sqdem", ignore.stderr=TRUE)
Check that it imported into GRASS ok:
> execGRASS("r.info", parameters=list(map="sqdemSP"))
+----------------------------------------------------------------------------+ | Layer: sqdemSP Date: Sun May 14 21:59:26 2006 | | Mapset: rsb Login of Creator: rsb | | Location: spearfish57 | | DataBase: /home/rsb/topics/grassdata | | Title: ( sqdemSP ) | |----------------------------------------------------------------------------| | | | Type of Map: raster Number of Categories: 255 | | Data Type: FCELL | | Rows: 477 | | Columns: 634 | | Total Cells: 302418 | | Projection: UTM (zone 13) | | N: 4928010 S: 4913700 Res: 30 | | E: 609000 W: 589980 Res: 30 | | Range of data: min = 32.649654 max = 42.895222 | | | | Data Source: | | | | | | | | Data Description: | | generated by r.in.gdal | | | | | +----------------------------------------------------------------------------+
v.krige usage example
v.krige is a GRASS python script (Addon) which performs kriging operations in the GRASS environment, using R functions for the back-end interpolation. It is present in GRASS 6.5svn, and further developed in GRASS 7svn. It requires a number of dependencies: python-rpy2 (needs to be "Rpy2", "Rpy" will not do unless it is rpy 2.x), then the following R-CRAN packages:
- gstat, spgrass6 (as above)
install.packages(c("gstat","spgrass6"))
- maptools
install.packages("maptools")
- automap (optional), with gpclib (or rgeos)
install.packages("automap")
install.packages("rgeos")