MB-System: Difference between revisions
m (→Export to GMT) |
|||
| (28 intermediate revisions by one other user not shown) | |||
| Line 1: | Line 1: | ||
* go to the [[Marine Science]] wiki page | * go to the [[Marine Science]] wiki page | ||
* go to the [[GRASS and GMT]] wiki page | * go to the [[GRASS and GMT]] wiki page | ||
* go to the [[LIDAR]] wiki page | |||
== MB-System == | == MB-System == | ||
| Line 11: | Line 13: | ||
: See the [[GRASS and GMT]] wiki help page for more information. | : See the [[GRASS and GMT]] wiki help page for more information. | ||
* Ungridded data points may be piped directly from | * Ungridded data points may be piped directly from [http://www.mbari.org/data/mbsystem/html/mblist.html mblist] to GRASS's {{cmd|v.in.ascii}} module. | ||
: {{cmd|d.vect}}'s zcolor= option can be used to color by 3D depth value. | : {{cmd|d.vect}}'s zcolor= option can be used to color by 3D depth value. | ||
: Use the {{cmd|v.colors}} module for colorizing point data in GRASS (''v.colors'' may be unsuitable for massive datasets). | : Use the {{cmd|v.colors}} module for colorizing point data in GRASS (''v.colors'' may be unsuitable for massive datasets). | ||
| Line 32: | Line 34: | ||
* Export Lat/Lon + depth data from XTF datafile into a GRASS Lat/Lon location | * Export Lat/Lon + depth data from XTF datafile into a GRASS Lat/Lon location | ||
<source lang="bash"> | |||
mblist -I 074.XTF -OXYz | v.in.ascii out=track074 x=1 y=2 fs=tab | mblist -I 074.XTF -OXYz | v.in.ascii out=track074 x=1 y=2 fs=tab | ||
</source> | |||
* Export Lat/Lon from the XTF datafile, reproject into the current GRASS location's projection, and import into GRASS with {{cmd|v.in.ascii}} | * Export Lat/Lon from the XTF datafile, reproject into the current GRASS location's projection with {{cmd|m.proj}}, and import into GRASS with {{cmd|v.in.ascii}} | ||
<source lang="bash"> | |||
mblist -I 074.XTF -OXY | m.proj -i | cut -f1 -d' ' | \ | mblist -I 074.XTF -OXY | m.proj -i | cut -f1 -d' ' | \ | ||
v.in.ascii out=track074 x=1 y=2 fs=tab | |||
</source> | |||
: * ''Idea'': write a v.in.cdl script that will parse a NetCDF/CDL file and automatically set v.in.ascii's column= option with attribute table column names and SQL column types (INTEGER, FLOAT, VARCHAR(), DATE, etc.). | : * ''Idea'': write a v.in.cdl script that will parse a NetCDF/CDL file and automatically set v.in.ascii's column= option with attribute table column names and SQL column types (INTEGER, FLOAT, VARCHAR(), DATE, etc.). | ||
| Line 43: | Line 49: | ||
====== Into a gridded raster map ====== | ====== Into a gridded raster map ====== | ||
* Read a series of .fbt pre-processed bathymetry files, reproject into the current projection, and grid into raster maps. | * Read a series of <tt>.fbt</tt> pre-processed bathymetry files, reproject into the current projection (e.g. a local Transverse Mercator), and grid into raster maps. | ||
First create .fbt .fnv and .inf summary files | First create .fbt .fnv and .inf summary files | ||
| Line 66: | Line 72: | ||
mbm_plot -F-1 -I survey-datalist -N | mbm_plot -F-1 -I survey-datalist -N | ||
./survey-datalist.cmd | ./survey-datalist.cmd | ||
ps2ps survey-datalist.ps | |||
</source> | </source> | ||
Based on the above PostScript file set GRASS region by eye: | Based on the above PostScript file set GRASS region by eye, or with d.zoom + 'g.region -p -t': | ||
<source lang="bash"> | |||
LL: n=40:48N s=40:37N w=168:40E e=169:06E | |||
</source> | |||
Convert to local projection with {{cmd|m.proj}}: | Convert to local projection with {{cmd|m.proj}}: | ||
<source lang="bash"> | |||
echo "168d40E 40d37N | echo "168d40E 40d37N | ||
169d06E 40d48N" | m.proj -i | 169d06E 40d48N" | m.proj -i | ||
</source> | |||
Here's a script to do a bunch of them: | |||
<source lang="bash"> | |||
ZONES=" | |||
168:19:14E/168:22:33E/46:50:04S/46:47:41S | |||
168:13:07E/168:16:53E/46:50:20S/46:47:50S | |||
168:15:45E/168:17:52E/46:53:21S/46:51:25S | |||
168:18E/168:31:12E/46:40:17S/46:36:05S" | |||
for ZONE in $ZONES ; do | |||
W=`echo "$ZONE" | cut -f1 -d/ | sed -e 's/:/d/' -e "s/:/'/"` | |||
E=`echo "$ZONE" | cut -f2 -d/ | sed -e 's/:/d/' -e "s/:/'/"` | |||
S=`echo "$ZONE" | cut -f3 -d/ | sed -e 's/:/d/' -e "s/:/'/"` | |||
N=`echo "$ZONE" | cut -f4 -d/ | sed -e 's/:/d/' -e "s/:/'/"` | |||
echo -e "$W $S\n$E $N" | m.proj -i --quiet | cut -f1 -d' ' | |||
echo | |||
done | |||
</source> | |||
Plug those numbers into {{cmd|g.region}}: | Plug those numbers into {{cmd|g.region}}: | ||
<source lang="bash"> | |||
g.region s=4616845.02 n=4636836.33 w=520872.47 e=524862.14 | g.region s=4616845.02 n=4636836.33 w=520872.47 e=524862.14 | ||
</source> | |||
Set the grid cell size to 5m, and align to whole numbers, | Set the grid cell size to 5m, and align to whole numbers, | ||
and check that the rows x columns is reasonable (smaller than 40000x40000): | and check that the rows x columns is reasonable (smaller than 40000x40000): | ||
<source lang="bash"> | |||
g.region res=5 -a -p | g.region res=5 -a -p | ||
</source> | |||
Adjust resolution to smaller grid size if needed (1000x1000 is fine for a summary image). | Adjust resolution to smaller grid size if needed (1000x1000 rows,cols is fine for a summary image). | ||
<source lang="bash"> | |||
g.region res=10 -a -p | g.region res=10 -a -p | ||
</source> | |||
Scan, read, and reproject all <tt>.fbt</tt> data into a single x,y,z text file: | |||
[[File:SlopePoint bathy.jpg|350px|thumb|right|A raw datalist fed to [http://www.mbari.org/data/mbsystem/html/mblist.html mblist] then piped through '{{cmd|m.proj}} -i' and {{cmd|r.in.xyz}} to form a gridded DEM.]] | |||
[[File:SlopePoint bathy.jpg|350px|thumb|right|A datalist fed to [http://www.mbari.org/data/mbsystem/html/mblist.html mblist] then piped through '{{cmd|m.proj}} -i' and {{cmd|r.in.xyz}} to form a gridded DEM.]] | |||
: '' | : ''Note that this file can get very large, perhaps too large for a 32bit OS/filesystem. In these cases you can pipe directly into {{cmd|r.in.xyz}} so no file is written to disk.'' | ||
: ''note that -Rw/e/s/n is used to skip over out-of-region data. MB-System and GMT will accept DDD:MM:SS.SSSh format as well as decimal degrees, just like GRASS.'' | : ''note that -Rw/e/s/n is used to skip over out-of-region data. MB-System and GMT will accept DDD:MM:SS.SSSh format as well as decimal degrees, just like GRASS.'' | ||
: The following assumes the output is projected, for import into WGS84 lat/lon skip the cs2cs command or at least change "%.3f" to "%.8f". | : The following assumes the output is projected, for import into WGS84 lat/lon skip the {{cmd|m.proj}}/[http://proj.maptools.org/man_cs2cs.html cs2cs] command or at least change "%.3f" to "%.8f". | ||
<source lang="bash"> | <source lang="bash"> | ||
OUTPROJ="`g.proj -jf`" | OUTPROJ="`g.proj -jf`" | ||
| Line 135: | Line 167: | ||
Analyze "n" map for number of pings per square-meter. | Analyze "n" map for number of pings per square-meter. | ||
: no-data cells not randomly distributed so we set n=0 to NULL so we aren't biasing the data as much | : no-data cells not randomly distributed so we set n=0 to NULL so we aren't biasing the data as much | ||
<source lang="bash"> | |||
r.null zoom_fbt.n setnull=0 | r.null zoom_fbt.n setnull=0 | ||
r.univar zoom_fbt.n | r.univar zoom_fbt.n | ||
</source> | |||
"mean:" is number of pings per square-meter for cells which have data. | "mean:" is number of pings per square-meter for cells which have data. | ||
Now analyze bathymetry data: | Now analyze bathymetry data: | ||
<source lang="bash"> | |||
r.univar zoom_fbt.median | r.univar zoom_fbt.median | ||
</source> | |||
and set some nice colors: | and set some nice colors: | ||
<source lang="bash"> | |||
r.colors zoom_fbt.median color=bcyr | r.colors zoom_fbt.median color=bcyr | ||
</source> | |||
Finally display it: | Finally display it: | ||
<source lang="bash"> | |||
d.mon x0 | d.mon x0 | ||
d.rast zoom_fbt.median | d.rast zoom_fbt.median | ||
d.legend zoom_fbt.median | d.legend zoom_fbt.median | ||
d.barscale at=5,5 | d.barscale at=5,5 | ||
</source> | |||
====== Direct import to a raster map ====== | |||
* {{AddonCmd|r.in.mb}} is a modified version of {{cmd|r.in.xyz}} which will communicate directly with MB-System via the ''mbio'' library. | |||
: ''see also [http://blogs.tekmap.ns.ca/archives/category/gtk-mb-system GTK+ MB-System]'' | |||
===== Create vector maps from .fnv nav data ===== | ===== Create vector maps from .fnv nav data ===== | ||
| Line 156: | Line 198: | ||
====== Add bounding box ====== | ====== Add bounding box ====== | ||
* ({{AddonCmd|v.in.mbsys_fnv}} does this automatically) | |||
<source lang="bash"> | <source lang="bash"> | ||
MAP= | MAP=swath_MB2004_026_0801_XTF | ||
v.db.addcol $MAP column="north DOUBLE PRECISION, \ | v.db.addcol $MAP column="north DOUBLE PRECISION, \ | ||
| Line 170: | Line 213: | ||
====== Import loop and patching ====== | ====== Import loop and patching ====== | ||
change DB to | change backend DB to SQLite: | ||
db.connect driver=sqlite database='$GISDBASE/$LOCATION_NAME/$MAPSET/sqlite.db' | db.connect driver=sqlite database='$GISDBASE/$LOCATION_NAME/$MAPSET/sqlite.db' | ||
| Line 224: | Line 267: | ||
d.what.vect datalist_tracks -f | d.what.vect datalist_tracks -f | ||
====== Create a single coverage area ====== | ====== Create a single coverage area ====== | ||
Convert all-swath | * Convert all-swath patches into a single polygon: | ||
: (''fast & good enough method'') | : (''fast & good enough method'') | ||
This method generalizes the data by converting into a raster grid then back into a vector area. | This method generalizes the data by converting into a raster grid then back into a vector area. | ||
| Line 250: | Line 292: | ||
# flatten to a boolean coverage mask map | # flatten to a boolean coverage mask map | ||
r.mapcalc "swath_coverage = int(swath_coverage)" | |||
r.reclass in=swath_coverage out=swath_coverage.1 << EOF | r.reclass in=swath_coverage out=swath_coverage.1 << EOF | ||
1 thru 999999 = 1 | 1 thru 999999 = 1 | ||
| Line 258: | Line 301: | ||
r.to.vect -s -v in=swath_coverage.1 out=Swath_coverage feature=area | r.to.vect -s -v in=swath_coverage.1 out=Swath_coverage feature=area | ||
# remove tiny holes (single or a few cells) | # remove tiny holes (single or a few cells, use interactive query to pick the threshold area) | ||
v.clean in=Swath_coverage out=Swath_coverage_noHoles tool=rmarea thresh=10000 | v.clean in=Swath_coverage out=Swath_coverage_noHoles tool=rmarea thresh=10000 | ||
</source> | </source> | ||
| Line 266: | Line 309: | ||
Clean | * Clean combined-swaths from ''v.patch'' and merge them into a single polygon: | ||
: (''much slower but more exact | : (''this way is much slower than the ''v.to.rast'' method, but the results are more exact'') | ||
<source lang="bash"> | <source lang="bash"> | ||
v.db.addcol map=datalist_swaths column='cat1 integer' | v.db.addcol map=datalist_swaths column='cat1 integer' | ||
| Line 273: | Line 316: | ||
v.reclass in=datalist_swaths out=swaths1 column=cat1 | v.reclass in=datalist_swaths out=swaths1 column=cat1 | ||
v.db.dropcol map=datalist_swaths column=cat1 | v.db.dropcol map=datalist_swaths column=cat1 | ||
# this cleaning step may take a ''very'' long time | # this cleaning step may take a ''very'' long time in GRASS 6 | ||
v.clean tool=break in=swaths1 out=swaths2 | v.clean tool=break in=swaths1 out=swaths2 | ||
v.dissolve in=swaths2 out=datalist_swaths_coverage | v.dissolve in=swaths2 out=datalist_swaths_coverage | ||
g.remove vect=swaths1,swaths3 | g.remove vect=swaths1,swaths3 | ||
</source> | </source> | ||
===== SQL queries ===== | |||
Examples: | |||
<source lang="bash"> | |||
d.vect datalist_swaths type=area color=150:170:200 fcolor=200:220:250 | |||
d.vect datalist_tracks width=2 | |||
d.zoom | |||
d.what.vect datalist_tracks -f | |||
db.select sql="SELECT length_m FROM track_MB2004_016_1856_XTF \ | |||
UNION SELECT length_m FROM track_MB2004_016_2145_XTF \ | |||
UNION SELECT length_m FROM track_MB2004_005_1742_XTF" | |||
db.select sql="SELECT length_m, filename \ | |||
FROM datalist_tracks WHERE length_m < 500" | |||
</source> | |||
* ''Please add more examples if you come up with any interesting tricks'' | |||
=== Export to GMT === | === Export to GMT === | ||
Processed raster grids and vector areas, lines, and points can be exported to back to GMT for plotting. See the [[GRASS and GMT]] wiki page for more information. | Processed raster grids and vector areas, lines, and points can be exported to back to GMT for plotting. See the [[GRASS and GMT]] wiki page for more information. | ||
* {{cmd|g.region}} in GRASS 6.5+ supports the ''-t'' flag to output the current region in MB-System's (and GMT's) <tt>W/E/S/N</tt> region format. | |||
* All of MB-System, GMT, and GRASS support the D:M:Sh notation for latitude and longitude, as well as decimal degrees. | |||
[[Category: Geodata]] | |||
[[Category: Import]] | |||
Latest revision as of 15:06, 20 December 2012
- go to the Marine Science wiki page
- go to the GRASS and GMT wiki page
- go to the LIDAR wiki page
MB-System
- MB-System is Free software for the processing and display of swath and sidescan sonar data. It can handle both multibeam bathymetry and sidescan sonar image data.
Import into GRASS
- Gridded data can be loaded into GRASS either as a GMT NetCDF grid via the r.in.gdal module (see GDAL supported formats), old-style GMT grid using r.in.bin -h, or as an Arc ASCII grid via the r.in.arc module.
- See the GRASS and GMT wiki help page for more information.
- Ungridded data points may be piped directly from mblist to GRASS's v.in.ascii module.
- d.vect's zcolor= option can be used to color by 3D depth value.
- Use the v.colors module for colorizing point data in GRASS (v.colors may be unsuitable for massive datasets).
- ".fnv" navigation files can be imported with the v.in.mbsys_fnv addon module in a number of different ways:
- track: ship's track
- port_trk: port-side outward track
- stbd_trk: starboard-side outward track
- scanlines: lines perpendicular to direction of travel
- swath: coverage area
- track_pts: ship's track as points
- all_pts: ship's track, port, and stbd track points
Examples
Import x,y,z bathymetry into GRASS
Into a vector points map
- Export Lat/Lon + depth data from XTF datafile into a GRASS Lat/Lon location
mblist -I 074.XTF -OXYz | v.in.ascii out=track074 x=1 y=2 fs=tab
- Export Lat/Lon from the XTF datafile, reproject into the current GRASS location's projection with m.proj, and import into GRASS with v.in.ascii
mblist -I 074.XTF -OXY | m.proj -i | cut -f1 -d' ' | \
v.in.ascii out=track074 x=1 y=2 fs=tab
- * Idea: write a v.in.cdl script that will parse a NetCDF/CDL file and automatically set v.in.ascii's column= option with attribute table column names and SQL column types (INTEGER, FLOAT, VARCHAR(), DATE, etc.).
- Interpolate into a DEM with (for example) the v.surf.rst or the v.to.rast+r.surf.nnbathy modules.
Into a gridded raster map
- Read a series of .fbt pre-processed bathymetry files, reproject into the current projection (e.g. a local Transverse Mercator), and grid into raster maps.
First create .fbt .fnv and .inf summary files
find . | grep '.[xX][tT][fF]$' > tmplist
mbdatalist -F-1 -I tmplist > datalist-1
mbdatalist -F-1 -I datalist-1 -N
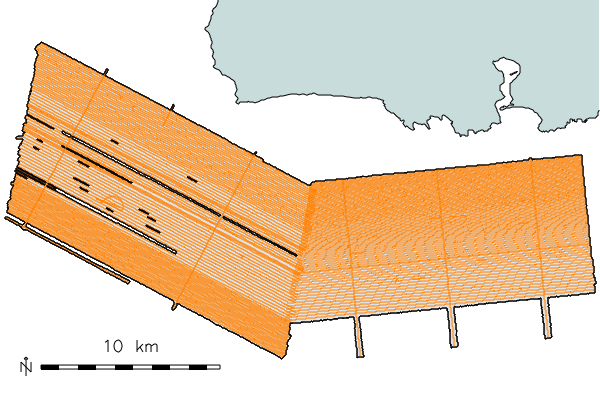
Next create quick GMT plot of the nav lines to check coverage
# scan multi-dir for bounds
for lltype in Longitude Latitude ; do
for tb in head tail ; do
grep $lltype `find | grep '\.inf$'` | \
awk '{print $3 "\n" $6}' | sort -n | $tb -n1
done
done
EXTENT=167.68/169.09/40.48/40.82
mbdatalist -F-1 -I datalist-1 -R$EXTENT > survey-datalist
mbm_plot -F-1 -I survey-datalist -N
./survey-datalist.cmd
ps2ps survey-datalist.ps
Based on the above PostScript file set GRASS region by eye, or with d.zoom + 'g.region -p -t':
LL: n=40:48N s=40:37N w=168:40E e=169:06E
Convert to local projection with m.proj:
echo "168d40E 40d37N
169d06E 40d48N" | m.proj -i
Here's a script to do a bunch of them:
ZONES="
168:19:14E/168:22:33E/46:50:04S/46:47:41S
168:13:07E/168:16:53E/46:50:20S/46:47:50S
168:15:45E/168:17:52E/46:53:21S/46:51:25S
168:18E/168:31:12E/46:40:17S/46:36:05S"
for ZONE in $ZONES ; do
W=`echo "$ZONE" | cut -f1 -d/ | sed -e 's/:/d/' -e "s/:/'/"`
E=`echo "$ZONE" | cut -f2 -d/ | sed -e 's/:/d/' -e "s/:/'/"`
S=`echo "$ZONE" | cut -f3 -d/ | sed -e 's/:/d/' -e "s/:/'/"`
N=`echo "$ZONE" | cut -f4 -d/ | sed -e 's/:/d/' -e "s/:/'/"`
echo -e "$W $S\n$E $N" | m.proj -i --quiet | cut -f1 -d' '
echo
done
Plug those numbers into g.region:
g.region s=4616845.02 n=4636836.33 w=520872.47 e=524862.14
Set the grid cell size to 5m, and align to whole numbers, and check that the rows x columns is reasonable (smaller than 40000x40000):
g.region res=5 -a -p
Adjust resolution to smaller grid size if needed (1000x1000 rows,cols is fine for a summary image).
g.region res=10 -a -p
Scan, read, and reproject all .fbt data into a single x,y,z text file:
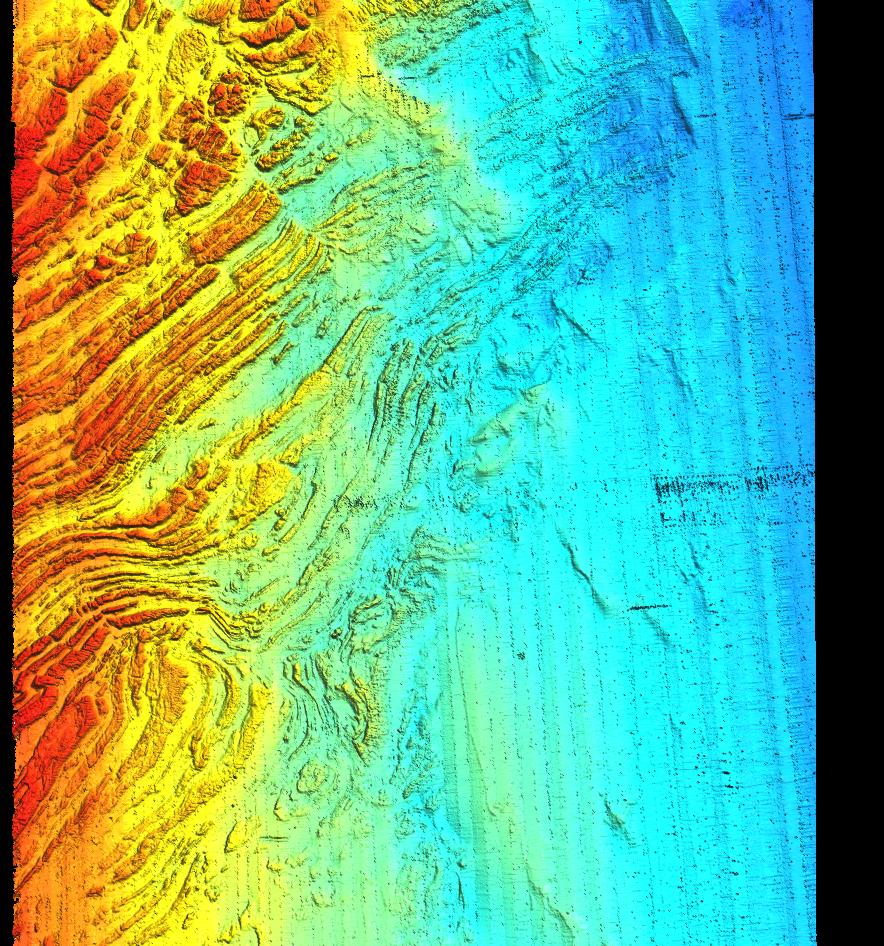
- Note that this file can get very large, perhaps too large for a 32bit OS/filesystem. In these cases you can pipe directly into r.in.xyz so no file is written to disk.
- note that -Rw/e/s/n is used to skip over out-of-region data. MB-System and GMT will accept DDD:MM:SS.SSSh format as well as decimal degrees, just like GRASS.
- The following assumes the output is projected, for import into WGS84 lat/lon skip the m.proj/cs2cs command or at least change "%.3f" to "%.8f".
OUTPROJ="`g.proj -jf`"
(
for FILE in `find . | grep '\.fbt$'` ; do
echo "Reading <$FILE> ..." 1>&2
mblist -I$FILE -D3 -R168:40E/169:06E/40:37N/40:48N
done
) | cs2cs -f '%.3f' +init=epsg:4326 +to $OUTPROJ | \
tr ' ' '\t' > fbt_UTM58_zoom.dat
Based on scan of the .inf files and personal knowledge set some reasonable bounds for the z-data:
# scan multi-dir for depth bounds
for tb in head tail ; do
grep Depth `find | grep '\.inf$'` | \
awk '{print $3 "\n" $6}' | sort -n | $tb -n1
done
#-2.7339
#200.6072
ZRANGE="-250,-2"
Run r.in.xyz to make aggregate raster maps.
r.in.xyz fs=tab x=1 x=2 z=3 zrange=$ZRANGE \
in=zoom_fbt.dat out=zoom_fbt.n method=n
r.in.xyz fs=tab x=1 x=2 z=3 zrange=$ZRANGE \
in=zoom_fbt.dat out=zoom_fbt.mean method=mean
r.in.xyz fs=tab x=1 x=2 z=3 zrange=$ZRANGE percent=25 \
in=zoom_fbt.dat out=zoom_fbt.median method=median
r.in.xyz fs=tab x=1 x=2 z=3 zrange=$ZRANGE percent=25 \
in=zoom_fbt.dat out=zoom_fbt.trim20 method=trimmean trim=20
Analyze "n" map for number of pings per square-meter.
- no-data cells not randomly distributed so we set n=0 to NULL so we aren't biasing the data as much
r.null zoom_fbt.n setnull=0
r.univar zoom_fbt.n
"mean:" is number of pings per square-meter for cells which have data.
Now analyze bathymetry data:
r.univar zoom_fbt.median
and set some nice colors:
r.colors zoom_fbt.median color=bcyr
Finally display it:
d.mon x0
d.rast zoom_fbt.median
d.legend zoom_fbt.median
d.barscale at=5,5
Direct import to a raster map
- r.in.mb is a modified version of r.in.xyz which will communicate directly with MB-System via the mbio library.
- see also GTK+ MB-System
Add bounding box
- (v.in.mbsys_fnv does this automatically)
MAP=swath_MB2004_026_0801_XTF
v.db.addcol $MAP column="north DOUBLE PRECISION, \
south DOUBLE PRECISION, east DOUBLE PRECISION, west DOUBLE PRECISION"
eval `v.info -g $MAP`
echo "UPDATE $MAP SET north = $north, south = $south, \
east = $east, west = $west" | db.execute
Import loop and patching
change backend DB to SQLite:
db.connect driver=sqlite database='$GISDBASE/$LOCATION_NAME/$MAPSET/sqlite.db'
Import all files in the datalist using a loop:
for TYPE in track swath ; do
for TRACK in `cut -f1 -d' ' datalist-1` ; do
OUTNAME="${TYPE}_$( echo "`basename $TRACK .fnv`" | sed -e 's/[- .]/_/g' )"
echo "Importing <$OUTNAME> ..."
CAT=`echo "$OUTNAME" | cut -f3,4 -d'_' | tr -d '_'`
v.in.mbsys_fnv in="${TRACK}.fnv" out="$OUTNAME" type=$TYPE cat=$CAT
done
done
Patch together all maps with a loop:
for TYPE in track swath ; do
# create an empty vector
v.in.ascii -e out="datalist_${TYPE}s" --quiet
# copy table structure
BASE_MAP=`g.mlist vect pattern="${TYPE}_*" | head -n 1`
exist_cols=`db.describe $BASE_MAP -c | grep '^Column' | cut -f2- -d: | \
sed -e 's/:/ /' -e 's/CHARACTER:\(.*\)/VARCHAR(\1)/' | \
cut -f1 -d: | tr '\n' ',' | sed -e 's/,$//' -e 's/^ //'`
v.db.addtable "datalist_${TYPE}s" columns="$exist_cols"
# run the patch loop
for MAP in `g.mlist vect pattern="${TYPE}_*"` ; do
echo "Patching <$MAP> ..."
v.patch -a -e in="$MAP" out=datalist_${TYPE}s --overwrite --quiet
done
done
- trac #859: v.patch is failing to preserve our carefully selected category numbers.
View and interactive query:
d.mon start=x0 d.vect datalist_swaths type=area color=150:170:200 fcolor=200:220:250 d.vect datalist_tracks width=2 d.zoom d.what.vect datalist_tracks -f
Create a single coverage area
- Convert all-swath patches into a single polygon:
- (fast & good enough method)
This method generalizes the data by converting into a raster grid then back into a vector area.
# display and set region & raster resolution
d.vect datalist_swaths type=area fcolor=none
d.zoom
g.region -a -p res=
# initialize base map
r.mapcalc "swath_coverage = 0"
# loop to systematically add a "1" every time a cell is
# covered by a swath; highlights overlap areas.
for MAP in `g.mlist vect pattern="swath_*"` ; do
echo "Adding <$MAP> ..."
v.to.rast in="$MAP" use=val val=1 type=area out=tmp.$MAP.$$ --quiet
r.series in=swath_coverage,tmp.$MAP.$$ method=sum out=tmp.series.$$ --quiet
g.remove tmp.$MAP.$$,swath_coverage --quiet
g.rename tmp.series.$$,swath_coverage --quiet
done
# flatten to a boolean coverage mask map
r.mapcalc "swath_coverage = int(swath_coverage)"
r.reclass in=swath_coverage out=swath_coverage.1 << EOF
1 thru 999999 = 1
0 = NULL
EOF
# convert back to a vector area polygon
r.to.vect -s -v in=swath_coverage.1 out=Swath_coverage feature=area
# remove tiny holes (single or a few cells, use interactive query to pick the threshold area)
v.clean in=Swath_coverage out=Swath_coverage_noHoles tool=rmarea thresh=10000
- Clean combined-swaths from v.patch and merge them into a single polygon:
- (this way is much slower than the v.to.rast method, but the results are more exact)
v.db.addcol map=datalist_swaths column='cat1 integer'
v.db.update map=datalist_swaths column=cat1 value=1
v.reclass in=datalist_swaths out=swaths1 column=cat1
v.db.dropcol map=datalist_swaths column=cat1
# this cleaning step may take a ''very'' long time in GRASS 6
v.clean tool=break in=swaths1 out=swaths2
v.dissolve in=swaths2 out=datalist_swaths_coverage
g.remove vect=swaths1,swaths3
SQL queries
Examples:
d.vect datalist_swaths type=area color=150:170:200 fcolor=200:220:250
d.vect datalist_tracks width=2
d.zoom
d.what.vect datalist_tracks -f
db.select sql="SELECT length_m FROM track_MB2004_016_1856_XTF \
UNION SELECT length_m FROM track_MB2004_016_2145_XTF \
UNION SELECT length_m FROM track_MB2004_005_1742_XTF"
db.select sql="SELECT length_m, filename \
FROM datalist_tracks WHERE length_m < 500"
- Please add more examples if you come up with any interesting tricks
Export to GMT
Processed raster grids and vector areas, lines, and points can be exported to back to GMT for plotting. See the GRASS and GMT wiki page for more information.
- g.region in GRASS 6.5+ supports the -t flag to output the current region in MB-System's (and GMT's) W/E/S/N region format.
- All of MB-System, GMT, and GRASS support the D:M:Sh notation for latitude and longitude, as well as decimal degrees.