R statistics/rgrass: Difference between revisions
Veroandreo (talk | contribs) m (→Querying maps) |
Veroandreo (talk | contribs) (→R within GRASS: setting proper format to chunks of R code) |
||
| Line 36: | Line 36: | ||
Load the ''rgrass7'' library: | Load the ''rgrass7'' library: | ||
<source lang="rsplus"> | |||
library(rgrass7) | |||
</source> | |||
If you plan to follow the example using the North Carolina sample dataset, load the ''rgdal'' library: | If you plan to follow the example using the North Carolina sample dataset, load the ''rgdal'' library: | ||
<source lang="rsplus"> | |||
library(rgdal) | |||
</source> | |||
Get the GRASS environment (mapset, region, map projection, etc.); you can display the metadata for your location by printing G: | Get the GRASS environment (mapset, region, map projection, etc.); you can display the metadata for your location by printing G: | ||
<source lang="rsplus"> | |||
G <- gmeta() | |||
gisdbase /home/neteler/grassdata | gisdbase /home/neteler/grassdata | ||
location nc_spm_08_grass7 | location nc_spm_08_grass7 | ||
| Line 58: | Line 62: | ||
+lat_0=33.75 +lon_0=-79 +x_0=609601.22 +y_0=0 +no_defs +a=6378137 | +lat_0=33.75 +lon_0=-79 +x_0=609601.22 +y_0=0 +no_defs +a=6378137 | ||
+rf=298.257222101 +towgs84=0.000,0.000,0.000 +to_meter=1 | +rf=298.257222101 +towgs84=0.000,0.000,0.000 +to_meter=1 | ||
</ | </source> | ||
=== Listing of existing maps === | === Listing of existing maps === | ||
List available vector maps: | List available vector maps: | ||
<source lang="rsplus"> | |||
execGRASS("g.list", parameters = list(type = "vector")) | |||
</source> | |||
List selected vector maps (wildcard): | List selected vector maps (wildcard): | ||
<source lang="rsplus"> | |||
execGRASS("g.list", parameters = list(type = "vector", pattern = "precip*")) | |||
</source> | |||
Save selected vector maps into R vector: | Save selected vector maps into R vector: | ||
<source lang="rsplus"> | |||
my_vmaps <- execGRASS("g.list", parameters = list(type = "vector", pattern = "precip*")) | |||
attributes(my_vmaps) | |||
attributes(my_vmaps)$resOut | |||
</source> | |||
List available raster maps: | List available raster maps: | ||
<source lang="rsplus"> | |||
execGRASS("g.list", parameters = list(type = "raster")) | |||
</source> | |||
List selected raster maps (wildcard): | List selected raster maps (wildcard): | ||
<source lang="rsplus"> | |||
execGRASS("g.list", parameters = list(type = "raster", pattern = "lsat7_2002*")) | |||
</source> | |||
=== Reading in data from GRASS === | === Reading in data from GRASS === | ||
| Line 87: | Line 101: | ||
# - geology_30m is a categorical map (i.e., it comes with classes) | # - geology_30m is a categorical map (i.e., it comes with classes) | ||
# - elevation is a continuous surface | # - elevation is a continuous surface | ||
<source lang="rsplus"> | |||
ncdata <- readRAST(c("geology_30m", "elevation"), cat=c(TRUE, FALSE)) | |||
</source> | |||
(A warning may appear since in the "geology_30m" map two labels are not unique - as found in the original data.) | (A warning may appear since in the "geology_30m" map two labels are not unique - as found in the original data.) | ||
We can verify the new R object: | We can verify the new R object: | ||
<source lang="rsplus"> | |||
str(ncdata) | |||
Formal class 'SpatialGridDataFrame' [package "sp"] with 4 slots | |||
..@ data :'data.frame': 16616 obs. of 2 variables: | |||
</source> | |||
and also check the data structure in more detail: | and also check the data structure in more detail: | ||
<source lang="rsplus"> | |||
str(ncdata@data) | |||
'data.frame': 16616 obs. of 2 variables: | |||
$ geology_30m: Factor w/ 12 levels "CZfg_217","CZlg_262",..: NA NA NA NA NA NA NA NA NA NA ... | |||
$ elevation : num NA NA NA NA NA NA NA NA NA NA ... | |||
</source> | |||
The metadata are now accessed and available, but are not (yet) used to structure the ''sp'' class objects, in this case a SpatialGridDataFrame object filled with data from two North Carolina layers. Here is a plot of the elevation data: | The metadata are now accessed and available, but are not (yet) used to structure the ''sp'' class objects, in this case a SpatialGridDataFrame object filled with data from two North Carolina layers. Here is a plot of the elevation data: | ||
<source lang="rsplus"> | |||
image(ncdata, "elevation", col = terrain.colors(20)) | |||
</source> | |||
Add a title to the plot: | Add a title to the plot: | ||
<source lang="rsplus"> | |||
title("North Carolina elevation") | |||
</source> | |||
[[Image:ncdata.png|center]] | [[Image:ncdata.png|center]] | ||
In addition, we can show what is going on inside the objects read into R: | In addition, we can show what is going on inside the objects read into R: | ||
<source lang="rsplus"> | |||
str(G) | |||
List of 26 | List of 26 | ||
$ DEBUG : chr "0" | $ DEBUG : chr "0" | ||
| Line 142: | Line 164: | ||
$ proj4 : chr "+proj=lcc +lat_1=36.16666666666666 +lat_2=34.33333333333334 +lat_0=33.75 +lon_0=-79 +x_0=609601.22 +y_0=0 +no_defs +a=6378137 +"| __truncated__ | $ proj4 : chr "+proj=lcc +lat_1=36.16666666666666 +lat_2=34.33333333333334 +lat_0=33.75 +lon_0=-79 +x_0=609601.22 +y_0=0 +no_defs +a=6378137 +"| __truncated__ | ||
- attr(*, "class")= chr "gmeta" | - attr(*, "class")= chr "gmeta" | ||
summary(ncdata) | |||
Object of class SpatialGridDataFrame | Object of class SpatialGridDataFrame | ||
Coordinates: | Coordinates: | ||
| Line 169: | Line 189: | ||
CZbg_910: 4351 Max. :156.25 | CZbg_910: 4351 Max. :156.25 | ||
(Other) : 4942 | (Other) : 4942 | ||
</ | </source> | ||
==== Example 2 ==== | ==== Example 2 ==== | ||
| Line 175: | Line 195: | ||
Here an example for a single map transfer from GRASS GIS to R: | Here an example for a single map transfer from GRASS GIS to R: | ||
<source lang=" | <source lang="rsplus"> | ||
library(rgrass7) | library(rgrass7) | ||
execGRASS("g.region", raster = "elevation", flags = "p") | execGRASS("g.region", raster = "elevation", flags = "p") | ||
| Line 186: | Line 206: | ||
We can create a table of cell counts: | We can create a table of cell counts: | ||
<source lang="rsplus"> | |||
table(ncdata$geology_30m) | |||
</source> | |||
{| class="wikitable" border="1" | {| class="wikitable" border="1" | ||
!CZfg_217 | !CZfg_217 | ||
| Line 218: | Line 240: | ||
And compare with the equivalent GRASS module: | And compare with the equivalent GRASS module: | ||
<source lang="rsplus"> | |||
execGRASS("r.stats", flags=c("c", "l"), parameters=list(input="geology_30m"), ignore.stderr=TRUE) | |||
</source> | |||
<pre> | <pre> | ||
217 CZfg 70381 | 217 CZfg 70381 | ||
| Line 236: | Line 260: | ||
Create a box plot of geologic types at different elevations: | Create a box plot of geologic types at different elevations: | ||
<source lang="rsplus"> | |||
boxplot(ncdata$elevation ~ ncdata$geology_30m, medlwd = 1) | |||
</source> | |||
[[Image:boxplot_geo.png|center]] | [[Image:boxplot_geo.png|center]] | ||
| Line 280: | Line 306: | ||
First prepare some data: (square root of elevation) | First prepare some data: (square root of elevation) | ||
<source lang="rsplus"> | |||
ncdata$sqdem <- sqrt(ncdata$elevation) | |||
</source> | |||
Export data from ''R'' back into a GRASS raster map: | Export data from ''R'' back into a GRASS raster map: | ||
<source lang="rsplus"> | |||
writeRAST(ncdata, "sqdemNC", zcol="sqdem", ignore.stderr=TRUE) | |||
</source> | |||
Check that it imported into GRASS ok: | Check that it imported into GRASS ok: | ||
<source lang="rsplus"> | |||
execGRASS("r.info", parameters=list(map="sqdemNC")) | |||
</source> | |||
<pre> | <pre> | ||
| Line 321: | Line 353: | ||
any analyses without loosing the possibility of still using GRASS command line, you can run: | any analyses without loosing the possibility of still using GRASS command line, you can run: | ||
< | <source lang="bash"> | ||
GRASS> rstudio & | GRASS> rstudio & | ||
</ | </source> | ||
or, if you already are working on a certain RStudio project: | or, if you already are working on a certain RStudio project: | ||
< | <source lang="bash"> | ||
GRASS> rstudio /path/to/project/folder/ & | GRASS> rstudio /path/to/project/folder/ & | ||
</ | </source> | ||
Then, you load rgrass7 library in your RStudio project | Then, you load rgrass7 library in your RStudio project | ||
<source lang="rsplus"> | |||
library(rgrass7) | |||
</source> | |||
and you are ready to go. | and you are ready to go. | ||
Revision as of 17:37, 15 September 2016
This page refers to the usage of R within a GRASS GIS 7 session and the use of GRASS GIS 7 within an R session. (see also R_statistics/spgrass6)
Terminology
Using R in conjunction with GRASS GIS can have two meanings:
- Using R within GRASS GIS session, i.e. you start R (or RStudio) from the GRASS GIS command line. You may like this variant if you are primarily a GIS user.
- Using GRASS GIS within a R session, i.e. you connect to a GRASS GIS location/mapset from within R (or RStudio). You may like this variant if you are primarily a R user.
References: see "Overview" in R_statistics
Installation
R within GRASS
Using R within GRASS GIS session, i.e. you start R (or RStudio) from the GRASS GIS command line.
Startup
- First start a GRASS GIS session. Then, at the GRASS command line start R (for a 'rstudio' session, see below)
- In this example we will use the North Carolina sample dataset.
Reset the region settings to the defaults
GRASS 7.0.1svn (nc_spm_08_grass7):~ > g.region -d
Launch R from the GRASS prompt
GRASS 7.0.1svn (nc_spm_08_grass7):~ > R
Load the rgrass7 library:
library(rgrass7)If you plan to follow the example using the North Carolina sample dataset, load the rgdal library:
library(rgdal)Get the GRASS environment (mapset, region, map projection, etc.); you can display the metadata for your location by printing G:
G <- gmeta()
gisdbase /home/neteler/grassdata
location nc_spm_08_grass7
mapset user1
rows 620
columns 1630
north 320000
south 10000
west 120000
east 935000
nsres 500
ewres 500
projection +proj=lcc +lat_1=36.16666666666666 +lat_2=34.33333333333334
+lat_0=33.75 +lon_0=-79 +x_0=609601.22 +y_0=0 +no_defs +a=6378137
+rf=298.257222101 +towgs84=0.000,0.000,0.000 +to_meter=1Listing of existing maps
List available vector maps:
execGRASS("g.list", parameters = list(type = "vector"))List selected vector maps (wildcard):
execGRASS("g.list", parameters = list(type = "vector", pattern = "precip*"))Save selected vector maps into R vector:
my_vmaps <- execGRASS("g.list", parameters = list(type = "vector", pattern = "precip*"))
attributes(my_vmaps)
attributes(my_vmaps)$resOutList available raster maps:
execGRASS("g.list", parameters = list(type = "raster"))List selected raster maps (wildcard):
execGRASS("g.list", parameters = list(type = "raster", pattern = "lsat7_2002*"))Reading in data from GRASS
Example 1
Read in two GRASS raster maps (the maps "geology_30m" and "elevation" from the North Carolina sample data location) into the R current session as a new object "ncdata" (containing then two SpatialGridDataFrames as well as metadata):
# the cat parameter indicates which raster values to be returned as factors # - geology_30m is a categorical map (i.e., it comes with classes) # - elevation is a continuous surface
ncdata <- readRAST(c("geology_30m", "elevation"), cat=c(TRUE, FALSE))(A warning may appear since in the "geology_30m" map two labels are not unique - as found in the original data.)
We can verify the new R object:
str(ncdata)
Formal class 'SpatialGridDataFrame' [package "sp"] with 4 slots
..@ data :'data.frame': 16616 obs. of 2 variables:and also check the data structure in more detail:
str(ncdata@data)
'data.frame': 16616 obs. of 2 variables:
$ geology_30m: Factor w/ 12 levels "CZfg_217","CZlg_262",..: NA NA NA NA NA NA NA NA NA NA ...
$ elevation : num NA NA NA NA NA NA NA NA NA NA ...The metadata are now accessed and available, but are not (yet) used to structure the sp class objects, in this case a SpatialGridDataFrame object filled with data from two North Carolina layers. Here is a plot of the elevation data:
image(ncdata, "elevation", col = terrain.colors(20))Add a title to the plot:
title("North Carolina elevation")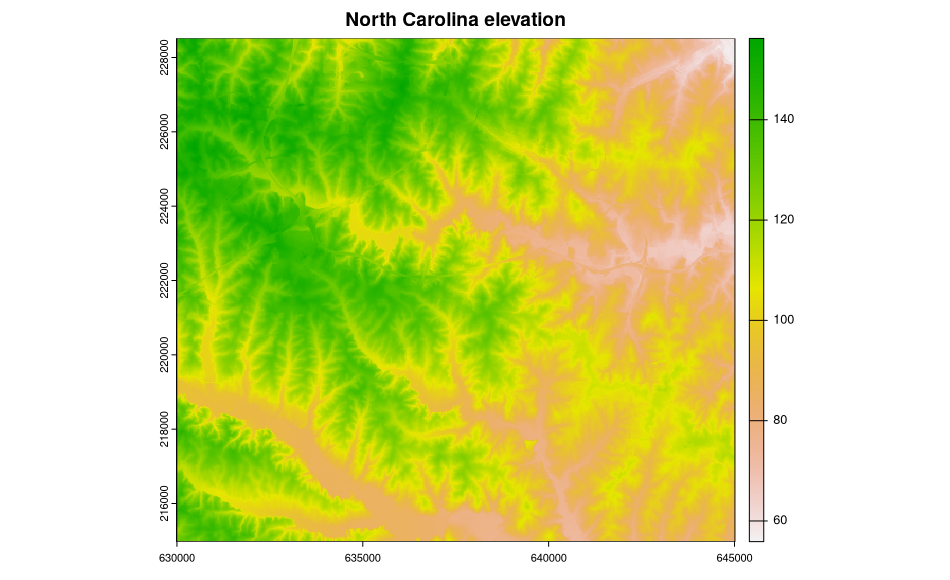
In addition, we can show what is going on inside the objects read into R:
str(G)
List of 26
$ DEBUG : chr "0"
$ LOCATION_NAME: chr "nc_spm_08_grass7"
$ GISDBASE : chr "/home/veroandreo/grassdata"
$ MAPSET : chr "PERMANENT"
$ GUI : chr "wxpython"
$ projection : chr "99"
$ zone : chr "0"
$ n : num 228500
$ s : num 215000
$ w : num 630000
$ e : num 645000
$ t : num 1
$ b : num 0
$ nsres : num 27.5
$ nsres3 : num 10
$ ewres : num 37.5
$ ewres3 : num 10
$ tbres : num 1
$ rows : int 491
$ rows3 : int 1350
$ cols : int 400
$ cols3 : int 1500
$ depths : int 1
$ cells : chr "196400"
$ cells3 : chr "2025000"
$ proj4 : chr "+proj=lcc +lat_1=36.16666666666666 +lat_2=34.33333333333334 +lat_0=33.75 +lon_0=-79 +x_0=609601.22 +y_0=0 +no_defs +a=6378137 +"| __truncated__
- attr(*, "class")= chr "gmeta"
summary(ncdata)
Object of class SpatialGridDataFrame
Coordinates:
min max
[1,] 630000 645000
[2,] 215000 228500
Is projected: TRUE
proj4string :
[+proj=lcc +lat_1=36.16666666666666 +lat_2=34.33333333333334
+lat_0=33.75 +lon_0=-79 +x_0=609601.22 +y_0=0 +no_defs +a=6378137
+rf=298.257222101 +towgs84=0.000,0.000,0.000 +to_meter=1]
Grid attributes:
cellcentre.offset cellsize cells.dim
1 630018.8 37.50000 400
2 215013.7 27.49491 491
Data attributes:
geology_30m elevation
CZfg_217:70381 Min. : 55.92
CZig_270:66861 1st Qu.: 94.78
CZbg_405:24561 Median :108.88
CZlg_262:19287 Mean :110.38
CZam_862: 6017 3rd Qu.:126.78
CZbg_910: 4351 Max. :156.25
(Other) : 4942Example 2
Here an example for a single map transfer from GRASS GIS to R:
library(rgrass7)
execGRASS("g.region", raster = "elevation", flags = "p")
ncdata <- readRAST("elevation", cat=FALSE)
summary(ncdata)
spplot(ncdata, col = terrain.colors(20))Summarizing data
We can create a table of cell counts:
table(ncdata$geology_30m)| CZfg_217 | CZlg_262 | CZig_270 | CZbg_405 | CZve_583 | CZam_720 | CZg_766 | CZam_862 | CZbg_910 | Km_921 | CZbg_945 | CZam_946 | CZam_948 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 70381 | 19287 | 66861 | 24561 | 2089 | 467 | 691 | 6017 | 4351 | 1211 | 1 | 398 | 85 |
And compare with the equivalent GRASS module:
execGRASS("r.stats", flags=c("c", "l"), parameters=list(input="geology_30m"), ignore.stderr=TRUE)217 CZfg 70381 262 CZlg 19287 270 CZig 66861 405 CZbg 24561 583 CZve 2089 720 CZam 467 766 CZg 691 862 CZam 6017 910 CZbg 4351 921 Km 1211 945 CZbg 1 946 CZam 398 948 CZam 85
Create a box plot of geologic types at different elevations:
boxplot(ncdata$elevation ~ ncdata$geology_30m, medlwd = 1)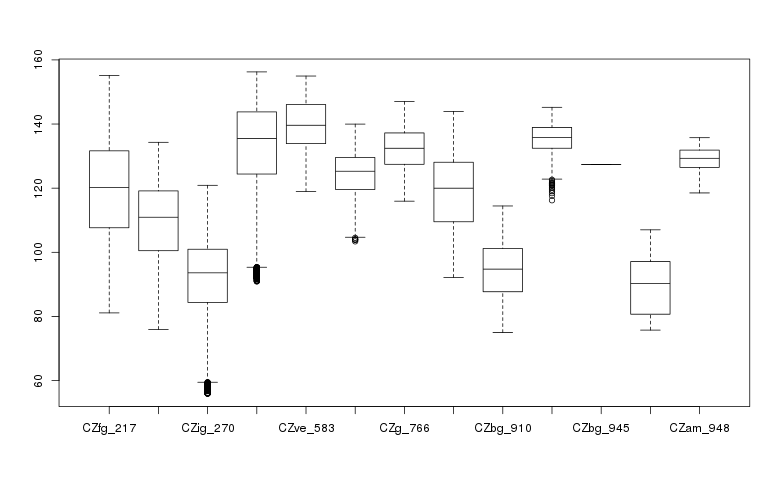
Querying maps
Sometimes you may want to query GRASS GIS maps from your R session.
As an example, here the transfer of raster pixel values at the position of vector points:
# set the computational region first to the raster map:
execGRASS("g.region", raster = "elev_state_500m", flags = "p")
# query raster map at vector points, transfer result into R
goutput <- execGRASS("r.what", map="elev_state_500m", points="geodetic_pts", separator=",", intern=TRUE)
str(goutput)
chr [1:29939] "571530.81289275,265739.968425953,,187.8082200648" ...
# parse it
con <- textConnection(goutput)
go1 <- read.csv(con, header=FALSE)
str(go1)
'data.frame': 29939 obs. of 4 variables:
$ V1: num 571531 571359 571976 572391 573011 ...
$ V2: num 265740 265987 267049 267513 269615 ...
$ V3: logi NA NA NA NA NA NA ...
$ V4: Factor w/ 22738 levels "-0.0048115728",..: 6859 6642 6749 6411 6356 6904 7506 7224 6908 7167 ...
summary(go1)
V1 V2 V3 V4
Min. :121862 Min. : 7991 Mode:logical 0 : 723
1st Qu.:462706 1st Qu.:162508 NA's:29939 * : 293
Median :610328 Median :204502 0.3048 : 47
Mean :588514 Mean :200038 0.6096 : 44
3rd Qu.:717610 3rd Qu.:247633 1.524 : 42
Max. :946743 Max. :327186 0.9144 : 23
(Other):28767Exporting data back to GRASS
Finally, a SpatialGridDataFrame object is written back to a GRASS raster map:
First prepare some data: (square root of elevation)
ncdata$sqdem <- sqrt(ncdata$elevation)Export data from R back into a GRASS raster map:
writeRAST(ncdata, "sqdemNC", zcol="sqdem", ignore.stderr=TRUE)Check that it imported into GRASS ok:
execGRASS("r.info", parameters=list(map="sqdemNC"))+----------------------------------------------------------------------------+ | Map: sqdemNC Date: Sun Jul 19 13:06:34 2015 | | Mapset: PERMANENT Login of Creator: veroandreo | | Location: nc_spm_08_grass7 | | DataBase: /home/veroandreo/grassdata | | Title: ( sqdemNC ) | | Timestamp: none | |----------------------------------------------------------------------------| | | | Type of Map: raster Number of Categories: 0 | | Data Type: DCELL | | Rows: 491 | | Columns: 400 | | Total Cells: 196400 | | Projection: Lambert Conformal Conic | | N: 228500 S: 215000.0002 Res: 27.49490794 | | E: 645000 W: 630000 Res: 37.5 | | Range of data: min = 7.47818253045085 max = 12.5000787351036 | | | | Data Description: | | generated by r.in.bin | | | | Comments: | | | +----------------------------------------------------------------------------+
Using RStudio in a GRASS GIS session
If you are most used to run R through RStudio, but still want to have all GRASS data available for performing any analyses without loosing the possibility of still using GRASS command line, you can run:
GRASS> rstudio &
or, if you already are working on a certain RStudio project:
GRASS> rstudio /path/to/project/folder/ &
Then, you load rgrass7 library in your RStudio project
library(rgrass7)and you are ready to go.
GRASS within R
Using GRASS GIS within a R session, i.e. you connect to a GRASS GIS location/mapset from within R (or RStudio).
Startup
In the first place, find out the path to the GRASS GIS library. This can be easily done with the following command (still outside of R; or through a system() call inside of R):
# OSGeo4W users: nothing to do # Linux, Mac OSX users: grass70 --config path
It may report something like:
/usr/local/grass-7.0.1
To call GRASS GIS 7 functionality from R, start R and use the initGRASS() function to define the GRASS settings:
## MS-Windows users:
library(rgrass7)
# initialisation and the use of North Carolina sample dataset
initGRASS(gisBase = "C:/OSGeo4W/apps/grass/grass-7.1.svn",
gisDbase = "C:/Users/marissa/GRASSdata/",
location = "nc_spm_08_grass7", mapset = "user1", SG="elevation")
## Linux, Mac OSX users:
library(rgrass7)
# initialisation and the use of North Carolina sample dataset
initGRASS(gisBase = "/usr/local/grass-7.0.1", home = tempdir(),
gisDbase = "/home/veroandreo/grassdata/",
location = "nc_spm_08_grass7", mapset = "user1", SG="elevation")
Note: the optional SG parameter is a 'SpatialGrid' object to define the ‘DEFAULT_WIND’ of the temporary location.
# set computational region to default (optional)
system("g.region -dp")
# verify metadata
gmeta()
# get two raster maps into R space
ncdata <- readRAST(c("geology_30m", "elevation"), cat=c(TRUE, FALSE))
# calculate object summaries
summary(ncdata$geology_30m)
CZfg_217 CZlg_262 CZig_270 CZbg_405 CZve_583 CZam_720 CZg_766 CZam_862
292 78 277 102 8 1 2 25
CZbg_910 Km_921 CZam_946 NA's
18 5 2 1009790
R in GRASS in batch mode
Run the following script with
R CMD BATCH batch.R
library(rgrass7)
# initialisation and the use of north carolina dataset
initGRASS(gisBase = "/home/veroandreo/software/grass-7.0.svn/dist.x86_64-unknown-linux-gnu",
home = tempdir(),
gisDbase = "/home/veroandreo/grassdata/",
location = "nc_spm_08_grass7", mapset = "user1", SG="elevation",
override = TRUE)
# set region to default
system("g.region -dp")
# verify
gmeta()
# read data into R
ncdata <- readRAST(c("geology_30m", "elevation"), cat=c(TRUE, FALSE))
# summary of geology map
summary(ncdata$geology_30m)
proc.time()
The result is (shorted here):
cat batch.Rout
R version 3.2.1 (2015-06-18) -- "World-Famous Astronaut"
Copyright (C) 2015 The R Foundation for Statistical Computing
Platform: x86_64-redhat-linux-gnu (64-bit)
...
> library(rgrass7)
Loading required package: sp
Loading required package: XML
GRASS GIS interface loaded with GRASS version: (GRASS not running)
> # initialisation and the use of north carolina dataset
> initGRASS(gisBase = "/home/veroandreo/software/grass-7.0.svn/dist.x86_64-unknown-linux-gnu", home = tempdir(),
+ gisDbase = "/home/veroandreo/grassdata/",
+ location = "nc_spm_08_grass7", mapset = "user1", SG="elevation",
+ override = TRUE)
gisdbase /home/veroandreo/grassdata/
location nc_spm_08_grass7
mapset user1
rows 620
columns 1630
north 320000
south 10000
west 120000
east 935000
nsres 500
ewres 500
projection +proj=lcc +lat_1=36.16666666666666 +lat_2=34.33333333333334
+lat_0=33.75 +lon_0=-79 +x_0=609601.22 +y_0=0 +no_defs +a=6378137
+rf=298.257222101 +towgs84=0.000,0.000,0.000 +to_meter=1
> system("g.region -dp")
projection: 99 (Lambert Conformal Conic)
zone: 0
datum: nad83
ellipsoid: a=6378137 es=0.006694380022900787
north: 320000
south: 10000
west: 120000
east: 935000
nsres: 500
ewres: 500
rows: 620
cols: 1630
cells: 1010600
> gmeta()
gisdbase /home/veroandreo/grassdata/
location nc_spm_08_grass7
mapset user1
rows 620
columns 1630
north 320000
south 10000
...
> ncdata <- readRAST(c("geology_30m", "elevation"), cat=c(TRUE, FALSE))
...
> summary(ncdata$geology_30m)
CZfg_217 CZlg_262 CZig_270 CZbg_405 CZve_583 CZam_720 CZg_766 CZam_862
292 78 277 102 8 1 2 25
CZbg_910 Km_921 CZam_946 NA's
18 5 2 1009790
> proc.time()
user system elapsed
8.061 2.016 10.048
Troubleshooting
Running out of disk space
Linux: A common issue is that /tmp/ is used for temporary files albeit being often a small partition. To change that to a larger directory, you may add to your $HOME/.bashrc the entry:
# change TMP directory of R (note: of course also another directory than suggested here is fine)
mkdir -p $HOME/tmp
export TMP=$HOME/tmp
The drawback is that on modern Linux systems the /tmp/ is a fast RAM disk (based on tempfs) while HOME directories are often on slower spinning disks (unless you have a SSD drive). At least you no longer run out of disk space easily.