R statistics
High quality statistic analysis in GRASS GIS is possible thanks to an interface to the most powerful statistics analysis package around: R (http://www.r-project.org).
Support for:
- The spgrass6 R addon package provides a convenient R ←→ GRASS GIS 6 interface
- The rgrass7 R addon package provides a convenient R ←→ GRASS GIS 7 interface
Using R in GRASS GIS directly can has two meanings:
- The first is that R is run "on top of" GRASS, transferring GRASS data to R to run statistical functions on the imported data as R objects in memory, and possibly transfer the results back to GRASS.
- The second is to leave the data mostly in GRASS, and to use R as a scripting language "on top of" GRASS with execGRASS() - in this case, little data is moved to R, so memory constraints are not important, but R functionality is available.
Quick start
For the impatient just start it:
> R
#and install packages directly from the net
pkgs <- c('akima', 'spgrass6', 'RODBC', 'VR', 'gstat')
install.packages(pkgs, dependencies=TRUE, type='source')
or to get all packages for spatial analysis in one:
> R
#To automatically install the spatial task view, the ctv package needs to be installed, e.g., via
install.packages("ctv")
library("ctv")
install.views("Spatial")
#or
update.views("Spatial")
Once you have R and spgrass6 on your system, have a look at this tutorial:
Installation
First of all you need to install R onto your system.
R and many of its addon packages are pre-built and distributed through the CRAN network of mirrors. In addition many Linux distributions prepackage R and a number of the most popular addon toolboxes.
All the necessary functions for the GRASS 6 interface are now in packages on CRAN, so that on Linux/Unix (or Mac OSX) installing rgdal from source with PROJ4 and GDAL installed, or Windows installing from binary, the required packages are: sp; maptools (now includes spmaptools); rgdal (now includes spGDAL, spproj); spgrass6 - now all on CRAN.
Status of "spgrass" and "rgrass7" packages
See overview here: https://r-forge.r-project.org/R/?group_id=2020
Source packages
From the R console first pick a local mirror:
chooseCRANmirror()
you can then see what it picked with
options("repos")
To permanently save the mirror site add it to ~/.Rprofile. For example:
options(repos=c(CRAN="http://cran.stat.auckland.ac.nz"))
and then run install.packages() as in the Quick Start section above.
For more information see http://cran.r-project.org/doc/manuals/R-admin.html
Linux
Debian and Ubuntu
R and a number of pre-build cran packages are already present in the main repositories. Start with:
# apt-get install r-base r-cran-vr r-cran-rodbc r-cran-xml
Once those are installed start "R" at the command prompt and install the libraries not packaged by the OS:
- n.b. r-cran-sp is now shipped as an official Debian/Ubuntu package and can be installed with apt as above
install.packages("sp")
install.packages("gstat")
Debian/Lenny ships with R 2.7.1 which is too old for the modern rgdal package. So we have to fetch an old one from the archive and build it from the Linux command line:
$ wget http://cran.r-project.org/src/contrib/Archive/rgdal/rgdal_0.6-24.tar.gz $ R CMD INSTALL -l /usr/local/lib/R/site-library rgdal_0.6-24.tar.gz
If you are using a newer version of R than that you can install the rgdal CRAN package directly:
install.packages("rgdal")
And finally, back inside the R session:
# in case of GRASS GIS 6
install.packages("spgrass6", dependencies = TRUE)
# in case of GRASS GIS 7
install.packages("rgrass7", dependencies = TRUE)
You can also use the CRAN Debian package repository: (pick one; adjust distribution as needed [here "Debian/testing"])
deb http://debian.cran.r-project.org/cran2deb/debian-i386 testing/ deb http://debian.cran.r-project.org/cran2deb/debian-amd64 testing/
RPM based
- RedHat, Fedora, openSuse, Mandriva and similar distros: take the latest R RPM and install it
R and a number of pre-build cran packages are already present in the main repositories. Start with:
# su # yum install R-core R-core-devel R-XML # exit
Once those are installed, start as normal user "R" at the command prompt and install the libraries not packaged by the distro provider:
R
# in case of GRASS GIS 6
install.packages("spgrass6", dependencies = TRUE)
# in case of GRASS GIS 7
install.packages("rgrass7", dependencies = TRUE)
Usage: You have the best user experience if you launch R within a running GRASS GIS session (then R automatically recognises the current settings of the Computational region and "sees" the GRASS maps).
Mac OSX
Start a R session, then
- for install.packages() you might have to rely on building packages from source code. try:
R
# in case of GRASS GIS 6
install.packages("spgrass6", type="source", dependencies = TRUE)
# in case of GRASS GIS 7
install.packages("rgrass7", type="source", dependencies = TRUE)
Startup of GRASS from within R:
First you need to find the path to the GRASS binaries: Control-click on the GRASS.app and you'll get a popup menu; select "Show Package Contents" - this opens you to the directory structure. Go to Contents->MacOS which would be "GISBASE"; So, in my case, the "gisBase" parameter is "/HD/Applications/Grass-6.4/Contents/MacOS". If you Command-click at the top of the window on the folder icon beside "MacOS" (from the line above this one), you can see the full path.
Now we can run GRASS from within a R session:
initGRASS(gisBase ='/Applications/GRASS/GRASS-6.4.app/Contents/MacOS',
location = 'geostat2012_ll', mapset = 'user1',
gisDbase = '/Users/Lars/Documents/Biologi/grassdata', override = TRUE)
Troubleshooting
If you get an error message when trying to call GRASS from R containing this line: dyld: Library not loaded: /usr/local/lib/libintl.8.dylib you need to establish a link from /Applications/Grass/GRASS-7.0.app/Contents/MacOS/lib/libintl.8.dylib to /usr/local/lib. This can be done through Terminal with the command:
sudo ln -s /Applications/Grass/GRASS-7.0.app/Contents/MacOS/lib/libintl.8.dylib /usr/local/lib/
Note: The path to the GRASS-x.x.app must reflect your own configuration.
MS Windows
Installation
Run:
# in case of GRASS GIS 6
install.packages("spgrass6", dependencies = TRUE)
# in case of GRASS GIS 7
install.packages("rgrass7", dependencies = TRUE)
or install Task View 'Spatial' - Analysis of Spatial Data
install.packages("ctv")
library("ctv")
install.views("Spatial")
GRASS 6: Calling R from GRASS
On Windows, the easiest way is calling R in a running GRASS 6 session by specifying the full path to the R startup executable. Note that all "\" must be changed to "/", furthermore quote white spaces in the path:
GRASS 6.4> C:/Users/"Catherine user"/Documents/R/R-2.15.1/bin/i386/R.exe R version 2.15.1 (2012-06-22) -- "Roasted Marshmallows" ... library(spgrass6)
In some cases, you can use this path:
- For 32 bits:
C:/"Program Files"/R/R-2.15.1/bin/i386/R.exe --save
- For 64 bits:
C:/"Program Files"/R/R-2.15.1/bin/x64/R.exe --save
The best solution is then to add this path to the %PATH% entry in the grass64.bat-starting script.
GRASS 6: calling GRASS from R
On Windows, the second easiest way is calling GRASS from R with initGRASS() from the R package spgrass6(). After GRASS has been initialized for R, you have access to GRASS commands from within R (you may need to adjust the path):
spgrass6()
loc <- initGRASS("C:\\Program Files\\GRASS 6.4", home=tempdir())
or using an already existing mapset:
spgrass6()
loc <- initGRASS(gisBase="C:\\Program Files\\GRASS 6.4",
gisDbase = "'C:\\Users\\neteler\\Documents\\grassdata'",
location = "spearfish60", mapset = "user1", SG="elevation.dem",
override = TRUE)
For an example, see http://geomorphometry.org/content/geomorphometry-r-saga-ilwis-grass
GRASS 6 Usage III (taken from the grass-stats-ML)
http://lists.osgeo.org/pipermail/grass-stats/2010-September/001274.html
at the moment there is following for a Grass-R-connection implemented in WinGRASS 64
(1) the WinGrass64-installer searches during installation for a installed R and writes - if found - the R-installation-path to %PATH% in the grass64.bat-starting script (see http://trac.osgeo.org/grass/browser/grass/branches/develbranch_6/mswindows/GRASS-Installer.nsi#L660)
--> this means that you FIRST have to install R and then winGRASS!
(2) you can start "GRASS-command-line" - it's a windows-command-line, not a msys-rxvt-terminal (you can find this starting option under Programs -> GRASS64 -> Grass command line; but not as a desktop icon)
with this starting option you start a Grass-session in the good old text mode. if you type R in the command line, then you start R inside a GRASS-session like in Linux.
GRASS 7 Usage
In WinGRASS 7 (standalone installer) the Windows batchfiles for use with R are now integrated for a smooth WinGRASS-R-integration. The R-installation-path is dynamically loaded into PATH.
Following R starting options are availabe:
- R cd - cd to R_ROOT, typically to C:\Program Files\R (0)
- R cmd - Run Rcmd.exe
- R dir - List contents of R_ROOT in chronological order showing R versions (0)
- R gui - Run Rgui.exe
- R help - Help info (0)
- R path - Add R_TOOLS, R_MIKTEX & R_PATH to path for this cmd line session (0)
- R Run R.exe (0)
- R script - Run Rscript.exe
- R show - Show R_ variable values used. R_PATH, etc. (0)
- R SetReg - Run RSetReg; see 2.17 in R FAQ for Windows (A)
- R tools - Add R_TOOLS and R_MIKTEX to path for this cmd line session (0)
- R touch - Change date on R_HOME to now (0) (A)
Start WinGRASS 7, bring the WinGRASS-windows console in front and type R for opening a R-session (command line mode) inside a GRASS-session.
Start WinGRASS 7, bring the WinGRASS-windows console in front and type R gui for opening a R-session (GUI mode) inside a GRASS-session.
Open tickets
- Ticket trac #1103 (new enhancement) WinGrass64 - windows-commandline not released: a Grass-session with wxGui, command-line and R inside a Grass-session would be possible (as already does in WinGrass7)
Command help
Start the R help browser:
help.start()
- Select Packages and then spgrass6.
Running
- by Roger Bivand
The R interface for GRASS 5.4 was provided by a CRAN package called grass. Changes going forward to the current GRASS 6 release meant that the interface had to be rewritten, and this provided the opportunity to adapt it to the sp CRAN package classes. Because GRASS provides the same kinds of data as sp classes handle, and relies on much of the same open source infrastructure (PROJ.4, GDAL, OGR), this step seemed sensible. Wherever possible spgrass6 tries to respect the current region in GRASS to avoid handling raster data with different resolutions or extents. R is assumed to be running within GRASS:
Startup
- Start GRASS. At the GRASS command line start R.
- In this example we will use the sample Spearfish dataset.
Reset the region settings to the defaults
GRASS> g.region -d
Launch R from the GRASS prompt
GRASS> R
Load the spgrass6 library:
> library(spgrass6)
Get the GRASS environment (mapset, region, map projection, etc.); you can display the metadata for your location by printing G:
> G <- gmeta6()
Listing of existing maps
List available vector maps:
execGRASS("g.list", parameters = list(type = "vect"))
List selected vector maps (wildcard):
execGRASS("g.list", parameters = list(type = "vect", pattern = "precip*"))
Save selected vector maps into R vector:
my_vmaps <- execGRASS("g.list", parameters = list(type = "vect", pattern = "precip*"))
attributes(my_vmaps)
attributes(my_vmaps)$resOut
List available raster maps:
execGRASS("g.list", parameters = list(type = "rast"))
List selected raster maps (wildcard):
execGRASS("g.list", parameters = list(type = "rast", pattern = "lsat7_2000*"))
Reading in data
Read in two raster maps (Spearfish sample dataset):
> spear <- readRAST6(c("geology", "elevation.dem"),
cat=c(TRUE, FALSE), ignore.stderr=TRUE,
plugin=NULL)
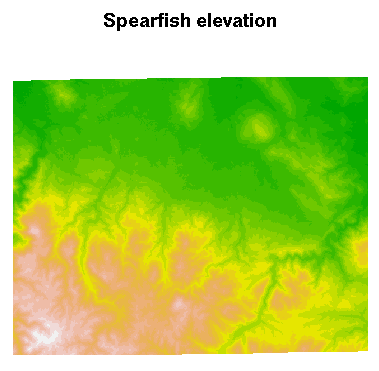
The metadata are accessed and available, but are not (yet) used to structure the sp class objects, here a SpatialGridDataFrame object filled with data from two Spearfish layers. Here is a plot of the elevation data:
> image(spear, attr = 2, col = terrain.colors(20))
Add a title to the plot:
> title("Spearfish elevation")
In addition, we can show what is going on inside the objects read into R:
> str(G)
List of 26 $ GISDBASE : chr "/home/rsb/topics/grassdata" $ LOCATION_NAME: chr "spearfish57" $ MAPSET : chr "rsb" $ DEBUG : chr "0" $ GRASS_GUI : chr "text" $ projection : chr "1 (UTM)" $ zone : chr "13" $ datum : chr "nad27" $ ellipsoid : chr "clark66" $ north : num 4928010 $ south : num 4913700 $ west : num 589980 $ east : num 609000 $ top : num 1 $ bottom : num 0 $ nsres : num 30 $ nsres3 : num 30 $ ewres : num 30 $ ewres3 : num 30 $ tbres : num 1 $ rows : int 477 $ rows3 : int 477 $ cols : int 634 $ cols3 : int 634 $ depths : int 1 $ proj4 : chr "+proj=utm +zone=13 +a=6378206.4 +rf=294.9786982 +no_defs +nadgrids=/home/rsb/topics/grass61/grass-6.1.cvs/etc/nad/conus"
> summary(spear)
Object of class SpatialGridDataFrame
Coordinates:
min max
coords.x1 589980 609000
coords.x2 4913700 4928010
Is projected: TRUE
proj4string : [+proj=utm +zone=13 +a=6378206.4 +rf=294.9786982 +no_defs +nadgrids=/home/rsb/topics/grass61/grass-6.1.cvs/etc/nad/conus]
Number of points: 2
Grid attributes:
cellcentre.offset cellsize cells.dim
1 589995 30 634
2 4913715 30 477
Data attributes:
geology elevation.dem
sandstone:74959 Min. : 1066
limestone:61355 1st Qu.: 1200
shale :46423 Median : 1316
sand :36561 Mean : 1354
igneous :36534 3rd Qu.: 1488
(Other) :37636 Max. : 1840
NA's : 8950 NA's :10101
Summarizing data
We can create a table of cell counts:
> table(spear$geology)
| metamorphic | transition | igneous | sandstone | limestone | shale | sandy shale | claysand | sand |
|---|---|---|---|---|---|---|---|---|
| 11693 | 142 | 36534 | 74959 | 61355 | 46423 | 11266 | 14535 | 36561 |
And compare with the equivalent GRASS module:
> execGRASS("r.stats", flags=c("c", "l"), parameters=list(input="geology"), ignore.stderr=TRUE)
1 metamorphic 11693 2 transition 142 3 igneous 36534 4 sandstone 74959 5 limestone 61355 6 shale 46423 7 sandy shale 11266 8 claysand 14535 9 sand 36561 * no data 8950
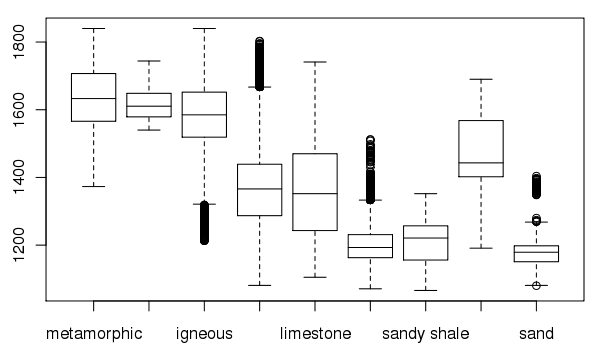
Create a box plot of geologic types at different elevations:
> boxplot(spear$elevation.dem ~ spear$geology, medlwd = 1)
Exporting data back to GRASS
Finally, a SpatialGridDataFrame object is written back to a GRASS raster map:
First prepare some data: (square root of elevation)
> spear$sqdem <- sqrt(spear$elevation.dem)
Export data from R back into a GRASS raster map:
> writeRAST6(spear, "sqdemSP", zcol="sqdem", ignore.stderr=TRUE)
Check that it imported into GRASS ok:
> execGRASS("r.info", parameters=list(map="sqdemSP"))
+----------------------------------------------------------------------------+ | Layer: sqdemSP Date: Sun May 14 21:59:26 2006 | | Mapset: rsb Login of Creator: rsb | | Location: spearfish57 | | DataBase: /home/rsb/topics/grassdata | | Title: ( sqdemSP ) | |----------------------------------------------------------------------------| | | | Type of Map: raster Number of Categories: 255 | | Data Type: FCELL | | Rows: 477 | | Columns: 634 | | Total Cells: 302418 | | Projection: UTM (zone 13) | | N: 4928010 S: 4913700 Res: 30 | | E: 609000 W: 589980 Res: 30 | | Range of data: min = 32.649654 max = 42.895222 | | | | Data Source: | | | | | | | | Data Description: | | generated by r.in.gdal | | | | | +----------------------------------------------------------------------------+
Calling GRASS functionality in R batch job
To call GRASS functionality within a R batch job, use the initGRASS() function to define the GRASS settings:
library(spgrass6)
# initialisation and the use of spearfish60 data
initGRASS(gisBase = "/usr/local/grass-6.4.1", home = tempdir(),
gisDbase = "/home/neteler/grassdata/",
location = "spearfish60", mapset = "user1", SG="elevation.dem",
override = TRUE)
system("g.region -d")
# verify
gmeta6()
spear <- readRAST6(c("geology", "elevation.dem"),
cat=c(TRUE, FALSE), ignore.stderr=TRUE,
plugin=NULL)
summary(spear$geology)
Run this script with
R CMD BATCH batch.R
The result is (shorted here):
cat batch.Rout
R version 2.10.0 (2009-10-26)
Copyright (C) 2009 The R Foundation for Statistical Computing
ISBN 3-900051-07-0
...
> library(spgrass6)
Loading required package: sp
Loading required package: rgdal
Geospatial Data Abstraction Library extensions to R successfully loaded
Loaded GDAL runtime: GDAL 1.7.2, released 2010/04/23
Path to GDAL shared files: /usr/local/share/gdal
Loaded PROJ.4 runtime: Rel. 4.7.1, 23 September 2009
Path to PROJ.4 shared files: (autodetected)
Loading required package: XML
GRASS GIS interface loaded with GRASS version: (GRASS not running)
>
> # initialisation and the use of spearfish60 data
> initGRASS(gisBase = "/usr/local/grass-6.4.1", home = tempdir(), gisDbase = "/home/neteler/grassdata/",
+ location = "spearfish60", mapset = "user1", SG="elevation.dem", override = TRUE)
gisdbase /home/neteler/grassdata/
location spearfish60
mapset user1
rows 477
columns 634
north 4928010
south 4913700
west 589980
east 609000
nsres 30
ewres 30
projection +proj=utm +zone=13 +a=6378206.4 +rf=294.9786982 +no_defs
+nadgrids=/usr/local/grass-6.4.1/etc/nad/conus +to_meter=1.0
Warning messages:
1: In dir.create(gisDbase) : '/home/neteler/grassdata' already exists
2: In dir.create(loc_path) :
'/home/neteler/grassdata//spearfish60' already exists
>
> system("g.region -d")
> # verify
> gmeta6()
gisdbase /home/neteler/grassdata/
location spearfish60
mapset user1
rows 477
columns 634
north 4928010
...
>
> spear <- readRAST6(c("geology", "elevation.dem"),
+ cat=c(TRUE, FALSE), ignore.stderr=TRUE,
+ plugin=NULL)
>
> summary(spear$geology)
metamorphic transition igneous sandstone limestone shale
11693 142 36534 74959 61355 46423
sandy shale claysand sand NA's
11266 14535 36561 8950
>
>
> proc.time()
user system elapsed
2.891 0.492 3.412
GRASS Modules
v.krige
v.krige is a GRASS python script which performs kriging operations in the GRASS environment, using R functions for the back-end interpolation. It is present in GRASS 6.5svn, and further developed in GRASS 7svn. It requires a number of dependencies: python-rpy2 (needs to be "Rpy2", "Rpy" will not do unless it is rpy 2.x), then the following R-CRAN packages:
- gstat, spgrass6 (as above)
install.packages(c("gstat","spgrass6"))
- maptools
install.packages("maptools")
- automap (optional), with gpclib (or rgeos)
install.packages("automap")
install.packages("rgeos")
Getting Support
- Primary support for R + GRASS and the spgrass6 package is through the grass-stats mailing list.
See also
- R. Bivand, 2007: Interfacing R and OSGeo projects: status and perspectives (Presentation with slides and scripts)
- Using GRASS and R: http://grassold.osgeo.org/statsgrass/grass6_r_interface.html
- Connecting R to RDBMS: http://grassold.osgeo.org/statsgrass/r_and_dbms.html
- Neural Networks with GRASS and R (posted by Markus Neteler on the grass-user mailing list) http://www.uam.es/proyectosinv/Mclim/pdf/MBenito_EcoMod.pdf
- A detailed example on the use of GRASS and R, with spearfish data: http://casoilresource.lawr.ucdavis.edu/drupal/node/438
- Using R and GRASS with cygwin: It is possible to use Rterm inside the GRASS shell in cygwin, just as in Unix/Linux or OSX. You should not, however, start Rterm from a cygwin xterm, because Rterm is not expecting to be run in an xterm under Windows, and loses its input. If you use the regular cygwin bash shell, but need to start display windows, start X from within GRASS with startx &, and then start Rterm in the same cygwin shell, not in the xterm.
- Spatial data in R (
sp) is a R library that provides classes and methods for spatial data (points, lines, polygons, grids), and to new or existing spatial statistics R packages that use sp, depend on sp, or will become dependent onsp, such asmaptools,rgdal,splancs,spgrass6,gstat,spgwrand many others.
- RPy - Python interface to the R Programming Language
Workshop material
- M. Neteler and M. Metz, 2011: Introduction to GRASS GIS. GEOSTAT 2011 Landau. Download workshop material (includes a R session)
Articles
- GRASS News vol.3, June 2005 (R. Bivand. Interfacing GRASS 6 and R. GRASS Newsletter, 3:11-16, June 2005. ISSN 1614-8746).
- OSGeo Journal vol. 1 May 2007 (R. Bivand. Using the R— GRASS interface. OSGeo Journal, 1:31-33, May 2007. ISSN 1614-8746).
- GRASS Book, last chapter