LIDAR: Difference between revisions
m (→LIDAR Tools) |
(refer to other LiDAR workshops in this Wiki) |
||
| (96 intermediate revisions by 12 users not shown) | |||
| Line 1: | Line 1: | ||
[[Image:NagsHead.gif|right|thumb|300px|Animated LiDAR time series in GRASS GIS 7]] | |||
<h1> LIDAR and Multi-beam Swath bathymetry data </h1> | <h1> LIDAR and Multi-beam Swath bathymetry data </h1> | ||
Point cloud data, as a type of representation of 3D surfaces, are usually produced by airborne or on-ground laser scanning, also known as Light Detection and Ranging (LiDAR). The data are often provided as sets of very dense (x, y, z) points or in a more complex, public file binary format called LAS that may include multiple returns as well as intensities. GRASS GIS supports basic and advanced lidar data processing and analysis. | |||
== Modules == | == Modules == | ||
In this section various modules are introduced. | |||
=== Import === | === Import === | ||
* {{cmd|r.in.xyz}} - Create a raster map from an assemblage of many coordinates using univariate statistics. ([http://hamish.bowman.googlepages.com/grassfiles#xyz example]) | * {{cmd|r.in.xyz}} - Create a raster map from an assemblage of many coordinates using univariate statistics. ([http://hamish.bowman.googlepages.com/grassfiles#xyz example]) | ||
* {{cmd|r.in.lidar}} - (GRASS 7 only; GRASS must be compiled with libLAS support) Create a raster map from a binary LAS format LiDAR file (*.las) using univariate statistics and filtering. r.in.lidar is based on r.in.xyz. In addition to the options of r.in.xyz, r.in.lidar provides some basic lidar point filter options. | |||
* {{cmd|v.in.ascii}} - Import data from an ASCII file to GRASS vector format. | * {{cmd|v.in.ascii}} - Import data from an ASCII file to GRASS vector format. | ||
: ''Due to memory overhead vector point imports will be limited to a few million data points unless topology and database creation is skipped with the '''-bt''' flags''. It may also be useful to clip the import file to only accept points falling within the current region by using the '''-r''' flag. See {{cmd|g.region}} for details on specifying the region bounds. | : ''Due to memory overhead vector point imports will be limited to a few million data points unless topology and database creation is skipped with the '''-bt''' flags''. It may also be useful to clip the import file to only accept points falling within the current region by using the '''-r''' flag. See {{cmd|g.region}} for details on specifying the region bounds. | ||
* {{cmd|v.in.lidar}} - (GRASS 7 only; GRASS must be compiled with libLAS support). Creates a vector points file from a binary LAS format LiDAR file (*.las or *.laz). r.in.lidar also can create a new location based on the LAS file, and can filter the input points by return and subregion. | |||
=== Analysis === | === Analysis === | ||
| Line 14: | Line 24: | ||
* {{cmd|v.outlier}} - Removes outliers from vector point data. | * {{cmd|v.outlier}} - Removes outliers from vector point data. | ||
* {{cmd|v.lidar.edgedetection}} - | * {{cmd|v.lidar.edgedetection}} - Uses interpolation and edge detection to create a new vector points file of LiDAR data so that the resulting attribute table is reclassified with CAT=1 for points associated with the ground surface (i.e., terrain) and useful for interpolating a raster terrain (DEM) map, CAT=2 for points pertaining to edges of human-contructed objects, and CAT=3 for other points that could pertain to vegetation or other features. | ||
* {{cmd|v.lidar.growing}} - Building contour determination and | * {{cmd|v.lidar.growing}} - Building contour determination and region growing algorithm for determining the building inside. | ||
* {{cmd|v.lidar.correction}} - Correction of the v.lidar.growing output. It is the last of the three algorithms for LIDAR filtering. | * {{cmd|v.lidar.correction}} - Correction of the v.lidar.growing output. It is the last of the three algorithms for LIDAR filtering. | ||
''References:'' | |||
* Detailed description: [http://2006.foss4g.org/materialDisplayba74.html?contribId=48&sessionId=59&materialId=slides&confId=1 here] (FOSS4G 2006) | |||
* Summarised version: [http://www.sigte.udg.edu/jornadassiglibre2007/comun/2pdf/4.pdf LiDAR filtering with GRASS] (2007) | |||
* Calibration of the filtering parameters (around 20 parameters) by integrating the USGS UCODE and GRASS, see [http://www.isprs.org/proceedings/XXXVIII/1_4_7-W5/paper/Brovelli-126.pdf INVERSE CALIBRATION OF LIDAR FILTERING PARAMETERS: UCODE/GRASS INTEGRATION] | |||
* See also the [[#Related publications|Related publications section]] below | |||
=== Surface generation === | === Surface generation === | ||
| Line 29: | Line 46: | ||
* {{cmd|r.fillnulls}} - Fills no-data areas in raster maps using v.surf.rst splines interpolation. | * {{cmd|r.fillnulls}} - Fills no-data areas in raster maps using v.surf.rst splines interpolation. | ||
* {{AddonCmd|r.surf.nnbathy}} - Natural Neighbor interpolation using the 'nn' addon. | |||
== LAS support == | |||
The commands {{cmd|r.in.lidar}} and {{cmd|v.in.lidar}} offer LAS support. This requires that GRASS GIS is compiled with [http://www.liblas.org/ liblas] (optionally [http://www.laszip.org/ laszip]). | |||
<pre> | |||
# installation on Fedora/Centos/EPEL7 | |||
yum install liblas liblas-devel laszip | |||
# installation on Ubuntu | |||
apt-get install liblas-bin liblas-dev | |||
</pre> | |||
Configuration of GRASS GIS for libLAS support: | |||
<pre> | |||
./configure \ | |||
... | |||
--with-liblas | |||
... | |||
</pre> | |||
Then [[Compile and Install]]. | |||
== Swath Bathymetry Tools == | == Swath Bathymetry Tools == | ||
| Line 34: | Line 75: | ||
''see also the [[Marine_Science#Multibeam_sonar_processing]] wiki page'' | ''see also the [[Marine_Science#Multibeam_sonar_processing]] wiki page'' | ||
* The [http://david.p.finlayson.googlepages.com/swathwidth v. | * The [http://david.p.finlayson.googlepages.com/swathwidth v.swathwidth] module by David Finlayson for planning surveys. (development page) | ||
* An example of [http://bambi.otago.ac.nz/hamish/grass/gdal/sidescan_warp.html post-processing scanned paper sidescan swaths] using thin plate spline warping with [http://www.gdal.org/ GDAL's] "<tt>gdalwarp -tps</tt>" function. (debugging page) | * An example of [http://bambi.otago.ac.nz/hamish/grass/gdal/sidescan_warp.html post-processing scanned paper sidescan swaths] using thin plate spline warping with [http://www.gdal.org/ GDAL's] "<tt>gdalwarp -tps</tt>" function. (debugging page) | ||
* [[ | * [[MB-System|GRASS integration]] with [http://www.ldeo.columbia.edu/res/pi/MB-System/ MB-System] (GPL) software for processing Multibeam and Sidescan Sonar data. GRASS + [[MB-System|MBsys]] + [[GMT]] make a nice scriptable trio. | ||
== LIDAR Tools == | == LIDAR Tools == | ||
| Line 44: | Line 85: | ||
* {{cmd|r.terraflow}} - computation of flow direction, flow accumulation and other basic topographic terrain indices from massive raster digital elevation models (DEM). From the Duke University [http://terrain.cs.duke.edu/ STREAM] project. | * {{cmd|r.terraflow}} - computation of flow direction, flow accumulation and other basic topographic terrain indices from massive raster digital elevation models (DEM). From the Duke University [http://terrain.cs.duke.edu/ STREAM] project. | ||
* [http://mpa.itc.it/markus/grass61/demos/rlake/ Flood simulation] using {{cmd|r.lake}}. Includes fancy NVIZ visualization of Trento, Italy, by Markus Neteler. | * [http://mpa.itc.it/markus/grass61/demos/rlake/ Flood simulation] using {{cmd|r.lake}}. Includes fancy [[NVIZ]] visualization of Trento, Italy, by Markus Neteler. | ||
* [http://www.cs.unc.edu/~isenburg/software/ LAStools] are a set of simple command line tools | * [http://www.cs.unc.edu/~isenburg/software/ LAStools] are a set of simple command line tools for converting to/from ASCII, viewing, comparing, and compressing LIDAR data. ''While free to use source code is available for older verions, newer versions are not open source, only work on MS Windows, and are no longer free for commercial or government use.'' | ||
* [ | * [https://liblas.org/ libLAS] ASPRS LiDAR data translation tools | ||
* See also [[LIDAR#Analysis|LIDAR Analysis]] | * See also [[LIDAR#Analysis|LIDAR Analysis]] | ||
== Micro-tutorial == | == Micro-tutorial for LAS data import == | ||
''The following scripts are given for UNIX Bourne Shell; MS-Windows users should use the Msys terminal to use them.'' | |||
=== Preparation === | === Preparation === | ||
==== Conversion of text files to LAS ==== | ==== Conversion of text files to LAS ==== | ||
(''optional: While you do not need to do this conversion for GRASS import, the resulting files are much smaller than the uncompressed text files; additionally, they are in a defined format. On the other hand ASCII text files will survive decades without the need for external software.'') | |||
[http://www.liblas.org/ libLAS] supports the following column types: | [http://www.liblas.org/ libLAS] supports the following column types: | ||
| Line 92: | Line 137: | ||
Then convert: | Then convert: | ||
txt2las -parse trxyzis -i data.asc -o data.las | txt2las -parse trxyzis -i data.asc -o data.las | ||
=== Import === | |||
* For GRASS 6 use the [http://www.liblas.org libLAS] utilities to convert LAS data into ASCII text format for GRASS. | |||
* Data stored in text files (for example, .csv) can generally be imported directly into GRASS. | |||
* | * The data used in the rest of this micro-tutorial can be found at [http://www.appliedimagery.com/downloads/sampledata/Serpent%20Mound%20Model%20LAS%20Data.las appliedimagery.com]. | ||
==== Import ASCII text into a raster DEM ==== | |||
==== | ===== Check bounds of LAS file ===== | ||
Check bounds and SRS: | Check bounds and SRS: | ||
| Line 113: | Line 158: | ||
Spatial Reference +proj=utm +zone=17 +ellps=WGS84 +units=m | Spatial Reference +proj=utm +zone=17 +ellps=WGS84 +units=m | ||
After creating a suitable UTM zone 17 location (EPSG:32617) | ===== Check bounds of ASCII text file ===== | ||
set the region according to the information from lasinfo at 1m resolution, | |||
This is similar in form to the above, but use the '-s' scan flag with the {{cmd|r.in.xyz}} module. Add the '-g' flag to get output ready for {{Cmd|g.region}}. | |||
===== Set region bounds and grid size ===== | |||
After creating a suitable UTM zone 17 location (EPSG:32617; the {{wikipedia|Serpent_Mound}} is in Ohio, USA) | |||
set the region according to the information from lasinfo at 1m resolution, using the '-a' flag to round the grid outwards, aligning to whole meters: | |||
GRASS> {{cmd|g.region}} n=4323641.57 s=4320942.61 w=289020.90 e=290106.02 res=1 -ap | GRASS> {{cmd|g.region}} n=4323641.57 s=4320942.61 w=289020.90 e=290106.02 res=1 -ap | ||
===== Run the import ===== | |||
Finally, import with {{cmd|r.in.xyz}} with data piped directly from the <tt>las2txt</tt> program and set a nice equalized color table: | Finally, import with {{cmd|r.in.xyz}} with data piped directly from the <tt>las2txt</tt> program and set a nice equalized color table: | ||
| Line 121: | Line 174: | ||
las2txt --stdout "${BASEMAP}_Data.las" | \ | las2txt --stdout "${BASEMAP}_Data.las" | \ | ||
r.in.xyz in=- out=${BASEMAP}_Data fs= | r.in.xyz in=- out=${BASEMAP}_Data fs=comma method=mean | ||
{{cmd|r.colors}} ${BASEMAP}_Data color=bcyr -e | {{cmd|r.colors}} ${BASEMAP}_Data color=bcyr -e | ||
The above example uses the default z-elevation level as the 3rd term, but by using the <tt>las2txt --parse</tt> command other fields (such as intensity) can be imported instead via "<tt>--parse xyi</tt>". | * The above example uses the default z-elevation level as the 3rd term, but by using the <tt>las2txt --parse</tt> command other fields (such as intensity) can be imported instead via "<tt>--parse xyi</tt>". Unfortunately there seem to be a number of versions of las2txt and each are called slightly differently. You might have to experiment a little to get the right incantation. | ||
* To import from an ASCII text file, run ''r.in.xyz'' directly with <tt>input=</tt> set to the filename instead of "-" (which indicates input will be piped in from another program). | |||
==== Direct import of LAS as raster DEM ==== | |||
This is the same as the above, but use the {{Cmd|r.in.lidar}} module (only in GRASS 7). In all cases you should manually decide on a suitable grid resolution and bounds, then set them with ''g.region'', before running the import modules. | |||
==== Import as vector points ==== | ==== Import LAS as vector points in GRASS 6 ==== | ||
Region setting (establishing the grid) is not needed for vector features so we can go directly to the import step. To deal with millions of input points {{cmd|v.in.ascii}} should be run with the options to skip creation of an attribute database and building topology as these can consume large amounts of memory. Note that vector maps without topology built are somewhat limited in their ability to be processed. Most LIDAR specific modules have been adapted to not require | Region setting (establishing the grid) is not needed for vector features so we can go directly to the import step. To deal with millions of input points {{cmd|v.in.ascii}} should be run with the options to skip creation of an attribute database and building topology as these can consume large amounts of memory. Note that vector maps without topology built are somewhat limited in their ability to be processed. Most LIDAR specific modules have been adapted to not require | ||
| Line 133: | Line 192: | ||
las2txt --stdout "${BASEMAP}_Data.las" | \ | las2txt --stdout "${BASEMAP}_Data.las" | \ | ||
v.in.ascii -tbz z=3 out="${BASEMAP}_pts" fs= | v.in.ascii -tbz z=3 out="${BASEMAP}_pts" fs=comma | ||
If topology was built, you can use {{cmd|d.vect}}'s -z flag to colorize by elevation value. Without topology you can still colorize, but you need to use color rules based on absolute elevations, not percentage of scale. | If topology was built, you can use {{cmd|d.vect}}'s -z flag to colorize by elevation value. Without topology you can still colorize, but you need to use color rules based on absolute elevations, not percentage of scale. | ||
| Line 144: | Line 203: | ||
* Vector points can be interpolated into raster DEMs with a number of modules, including {{cmd|v.surf.rst}}, {{cmd|v.surf.bspline}}, and {{cmd|v.surf.idw}}. | * Vector points can be interpolated into raster DEMs with a number of modules, including {{cmd|v.surf.rst}}, {{cmd|v.surf.bspline}}, and {{cmd|v.surf.idw}}. | ||
=== Clean raster DEM === | ==== Import LAS in GRASS 7 ==== | ||
As vector points | |||
* GRASS 7 offers the {{cmd|v.in.lidar}} command | |||
As raster: map: | |||
* {{cmd|r.in.lidar}} command | |||
* {{AddonSrc|imagery|r.in.pdal|version=7}} | |||
=== Clean imported raster DEM === | |||
(fill holes) | (fill holes) | ||
<source lang="bash"> | |||
# convert to vector points | # convert to vector points | ||
r.to.vect -z feature=point in=${BASEMAP}_Data out=${BASEMAP}_pt | r.to.vect -z feature=point in=${BASEMAP}_Data out=${BASEMAP}_pt | ||
| Line 164: | Line 233: | ||
# set colors to something nice | # set colors to something nice | ||
r.colors ${BASEMAP}.filled color=bcyr -e | r.colors ${BASEMAP}.filled color=bcyr -e | ||
</source> | |||
Depending on your needs, {{cmd|r.fillnulls}}, {{cmd|v.surf.bspline}}, {{cmd|v.surf.idw}}, {{cmd|r.surf.idw}}, {{cmd|r.surf.idw2}}, or [[GRASS_AddOns#r.surf.nnbathy|r.surf.nnbathy]] may be faster than the {{cmd|v.surf.rst}} method. | Depending on your needs, {{cmd|r.fillnulls}}, {{cmd|v.surf.bspline}}, {{cmd|v.surf.idw}}, {{cmd|r.surf.idw}}, {{cmd|r.surf.idw2}}, or [[GRASS_AddOns#r.surf.nnbathy|r.surf.nnbathy]] may be faster than the {{cmd|v.surf.rst}} method. | ||
| Line 182: | Line 251: | ||
It is possible to show vector points in 3D, but millions of them may make the program run slow. Topology is required ({{cmd|v.build}}). Tick the "3D" box in the Visualize → Vector points dialog. | It is possible to show vector points in 3D, but millions of them may make the program run slow. Topology is required ({{cmd|v.build}}). Tick the "3D" box in the Visualize → Vector points dialog. | ||
=== Export === | === LAS Export === | ||
==== Export to LAS ==== | ==== Export to LAS ==== | ||
| Line 194: | Line 263: | ||
txt2las --parse xyz -i ${BASEMAP}_export.txt | txt2las --parse xyz -i ${BASEMAP}_export.txt | ||
== Sample data == | == Micro-tutorial for LIDAR data analysis == | ||
=== Simple analysis of single return data === | |||
==== Import from ASCII file ==== | |||
Data source: North Carolina sample data set (Lidar data from 2001, near Raleigh, North Carolina). Download from | |||
http://www.grassbook.org/ncexternal/ | |||
-> File: BE3720079200WC20020829m.txt 3.6M (lidar bare earth points [spm]) | |||
Scan input file for spatial extent. The -g flag shows the result in a convenient copy-paste format for {{cmd|g.region}}: | |||
r.in.xyz BE3720079200WC20020829m.txt out=dummy -s -g | |||
We use this output to set region, predefine 2m raster cells, and polish the odd coordinates with -a: | |||
g.region n=222504.439000 s=219456.442000 e=640081.274000 w=637033.274000 res=2 -a -p | |||
==== QUESTION 1: Are these Lidar data sufficiently dense? ==== | |||
We compute a raster map representing number of points per cell | |||
r.in.xyz BE3720079200WC20020829m.txt out=lid_792_binn2m method=n | |||
Look at the resulting raster map: | |||
d.mon x0 | |||
d.font Vera | |||
d.rast.leg lid_792_binn2m | |||
Report point distribution in percent: | |||
r.report lid_792_binn2m unit=p | |||
r.univar lid_792_binn2m | |||
# (consider running `r.null setnull=0` first) | |||
Reduce the resolution to 6m to get at least one point per cell: | |||
g.region res=6 -a -p | |||
r.in.xyz BE3720079200WC20020829m.txt out=lid_792_binn6m method=n | |||
d.rast.leg lid_792_binn6m | |||
# ... a few holes remain but that's probably acceptable. | |||
Compute raster maps representing mean elevation for each cell: | |||
r.in.xyz BE3720079200WC20020829m.txt out=lid_792_binmean6m \ | |||
method=mean | |||
d.rast.leg lid_792_binmean6m | |||
Compute range and variation: | |||
r.in.xyz BE3720079200WC20020829m.txt out=lid_792_binrange6m \ | |||
method=range | |||
d.rast.leg lid_792_binrange6m | |||
r.in.xyz BE3720079200WC20020829m.txt out=lid_792_binvar6m \ | |||
method=coeff_var | |||
d.rast.leg lid_792_binvar6m | |||
Overlay other GIS maps to map of mean elevation: | |||
d.rast.leg lid_792_binmean6m | |||
d.vect streets_wake | |||
d.vect lakes type=boundary | |||
d.vect streams | |||
Now we continue to work in a zoomed spatial subset of the area. | |||
Here we only import only points in the rural area, do not build topology, and use the z-coordinate for elevation: | |||
g.region rural_1m -p | |||
v.in.ascii -ztbr BE3720079200WC20020829m.txt \ | |||
out=elev_lidrural_bepts z=3 | |||
Clear the monitor, then show a black and white orthophoto: | |||
d.erase -f | |||
d.rast ortho_2001_t792_1m | |||
We now use two LiDAR vector point maps, which were prepared earlier: | |||
* elev_lid792_bepts: Rural area (in tile 792), bare earth lidar point cloud | |||
* elev_lidrural_mrpts: Rural area multiple return lidar point cloud | |||
Look at ground data: | |||
d.vect elev_lidrural_bepts size=2 color=red | |||
Look at "surface" data: | |||
d.vect elev_lidrural_mrpts size=1 color=green | |||
Visualize 3D LiDAR multi-return points: | |||
nviz elev_lid792_1m point=elev_lidrural_mrpts | |||
# Visualize -> Vector Points -> 3D points | |||
# -> Icon size 2.25 | |||
# --> GRASS book p. 253, fig. 3.15 (see also screenshot [http://www.grassbook.org/about_menu3rd.php here]) | |||
==== DEM/DSM separation the simple way by selection of Lidar returns ==== | |||
Background on lidar multiple returns: "''Lidar returns are discrete observations* recorded when a laser pulse is intercepted and reflected by targets. Multiple returns derive from one laser pulse intercepting multiple targets (e.g. a top of a tree, its branches, and the ground)''." ([https://gis.stackexchange.com/a/143000/687 source: stackexchange]). | |||
Find out where we have multiple returns: | |||
d.rast ortho_2001_t792_1m | |||
d.vect elev_lidrural_mrpts where="return=1" color=red size=2 | |||
d.vect elev_lidrural_mrpts where="return=2" color=green size=3 | |||
d.vect elev_lidrural_mrpts where="return=3" color=blue | |||
d.vect elev_lidrural_mrpts where="return=4" color=yellow | |||
DTM: extract last return(s): | |||
v.extract elev_lidrural_mrpts out=elev_lidrural_mrpts_first where="Return < 2" | |||
nviz elev_lid792_1m point=elev_lidrural_mrpts_first | |||
Interpolate to map and look at it: | |||
v.surf.rst elev_lidrural_mrpts_first layer=0 elev=elev_lidrural_mrpts_DTM | |||
nviz elev_lidrural_mrpts_DTM col=ortho_2001_t792_1m | |||
DSM: extract first return(s): | |||
v.extract elev_lidrural_mrpts out=elev_lidrural_mrpts_last where="Return > 2" | |||
nviz elev_lid792_1m point=elev_lidrural_mrpts_last | |||
Interpolate to map and look at it: | |||
v.surf.rst elev_lidrural_mrpts_first layer=0 elev=elev_lidrural_mrpts_DSM | |||
nviz elev_lidrural_mrpts_DSM col=ortho_2001_t792_1m | |||
==== DEM/DSM separation the more complex way ==== | |||
<font color="red">''TODO: verify order of first and last returns in below text''</font> | |||
General procedure: | |||
* Lidar point clouds (first and last return) are imported with {{cmd|v.in.ascii}} (-b flag to not build the topology). | |||
* Outlier detection is done with {{cmd|v.outlier}} on both first and last return data (NB currently a selection of </=4 for soe/son in {{cmd|v.outlier}} results in an error message). | |||
* Then, with {{cmd|v.lidar.edgedetection}}, edges are detected from last return data. | |||
* The building are generated by {{cmd|v.lidar.growing}} from detected edges. | |||
* The resulting data are post-processed with {{cmd|v.lidar.correction}}. | |||
* Finally, the DTM and DSM are generated with {{cmd|v.surf.bspline}} (DTM: uses the 'v.lidar.correction' output; DSM: uses last return output from outlier detection). | |||
* NB for {{cmd|v.outlier}}, {{cmd|v.lidar.edgedetection}} and {{cmd|v.surf.bspline}}, one spline steps equates to 1m. It is recommended as a starting point that the choice of spline step is roughly 3 or 4 times the planimetric resolution (potential grid resolution) of your data. Experiment from there to obtain better results. | |||
export GRASS_OVERWRITE=1 | |||
v.extract elev_lidrural_mrpts out=elev_lidfirst_pts \ | |||
where="return = 1" | |||
v.extract elev_lidrural_mrpts out=elev_lidlast_pts \ | |||
where="return >= 2" | |||
d.vect elev_lidfirst_pts col=red | |||
d.vect elev_lidlast_pts col=green | |||
Outlier detection and separation into two maps | |||
# 1st return | |||
v.outlier elev_lidfirst_pts output=elev_lidfirst_clean \ | |||
outlier=elev_lidfirst_outl | |||
d.erase | |||
d.vect elev_lidfirst_clean size=2 | |||
d.vect elev_lidfirst_outl color=red | |||
2nd return | |||
v.outlier elev_lidlast_pts output=elev_lidlast_clean \ | |||
outlier=elev_lidlast_outl | |||
d.erase | |||
d.vect elev_lidlast_clean size=2 | |||
d.vect elev_lidlast_outl color=red | |||
# -> no outliers visible | |||
Run an edge detection on cleaned last return: | |||
v.lidar.edgedetection elev_lidlast_clean \ | |||
out=elev_lidlast_edges | |||
Buildings/vegetation are generated from detected edges (bug: you may need to specify the mapset): | |||
v.lidar.growing elev_lidlast_edges@lidar out=elev_lidlast_grow \ | |||
first=elev_lidfirst_clean | |||
Compare: | |||
d.vect elev_lidlast_pts col=blue | |||
d.vect elev_lidlast_grow col=green | |||
Correction (this is applied twice): | |||
v.lidar.correction elev_lidlast_grow out=elev_lidlast_corr1 \ | |||
terrain=elev_lidlast_terr1 | |||
v.lidar.correction elev_lidlast_corr1 out=elev_lid_dsm \ | |||
terrain=elev_lid_dtm | |||
DEM and DSM are generated: | |||
# Estimation of lambda_i parameter with cross validation (watch for RMS!) | |||
# and note use of bicubic for DSM and bilinear for DTM here and below | |||
v.surf.bspline -c elev_lid_dsm sie=100 sin=100 method=bicubic | |||
v.surf.bspline -c elev_lid_dtm sie=100 sin=100 method=bilinear | |||
# From the cross-validation, we select lambda with minimal RMS error: | |||
# generate raster surfaces at 1m resolution | |||
v.surf.bspline elev_lid_dsm raster=lidar_dsm lambda=0.1 method=bicubic | |||
v.surf.bspline elev_lid_dtm raster=lidar_dtm lambda=0.0001 method=bilinear | |||
d.rast lidar_dsm | |||
d.rast lidar_dtm | |||
nviz lidar_dsm,lidar_dtm \ | |||
col=ortho_2001_t792_1m,ortho_2001_t792_1m | |||
...with the position slider you can visually separate DSM and DEM, increase z slider (Visualize -> Raster Surface -> Position). | |||
=== Reclassification === | |||
As vector points | |||
* {{cmd|v.reclass}} command | |||
As raster map: | |||
* {{cmd|r.reclass}} command | |||
== Sample LIDAR data == | |||
=== Widely used in GRASS tutorials === | === Widely used in GRASS tutorials === | ||
| Line 226: | Line 489: | ||
* South Tyrol - Download of DTMs (Homepage in German or Italian) <BR> http://www.provinz.bz.it/raumordnung/grundkarten/utm/default_d.htm | * South Tyrol - Download of DTMs (Homepage in German or Italian) <BR> http://www.provinz.bz.it/raumordnung/grundkarten/utm/default_d.htm | ||
* libLAS's sample file collection | |||
:http://liblas.org/samples/ | |||
* NASA's Laser Vegetation Imaging Sensor (a.k.a. the Land, Vegetation, and Ice Sensor) or "LVIS" | |||
:https://lvis.gsfc.nasa.gov | |||
== Tutorials == | |||
* [[Lidar Analysis of Vegetation Structure]] | |||
* [[Processing_lidar_and_UAV_point_clouds_in_GRASS_GIS_(workshop_at_FOSS4G_Boston_2017)|Processing lidar and UAV point clouds in GRASS GIS]] | |||
* [http://ncsu-geoforall-lab.github.io/geospatial-modeling-course/topics/geomorphometry_basics.html Lectures and assignments from NCSU partially dealing with lidar] | |||
* [https://wenzeslaus.github.com/grass-lidar-talks Some basic and some new (7.2-7.4) GRASS GIS features related to lidar] | |||
;Breaklines | |||
* Not very well developed in GRASS so far, but we'd like to change that (you can volunteer) | |||
* {{AddonCmd|v.surf.icw}} (only suitable for ~ 200 input points) | |||
* [http://surfit.sourceforge.net/ SurfIt] (GPL, Tck/Tk) | |||
* <strike>[http://surgeweb.sweb.cz/surgemain.htm SurGe] (Trial/shareware) </strike> | |||
* [http://grasswiki.osgeo.org/wiki/TIN_with_breaklines v.triangle] add-on module for construction TIN with breaklines. | |||
: ''see above [[Contour lines to DEM]] page for further discussion of the (in)appropriateness of using TINs to generate raster surfaces. It is hoped that in future GRASS's more advanced spline interpolation modules will get breakline support.'' | |||
== Links == | == Links == | ||
* [[Contour lines to DEM]] interpolation module trials | |||
* [http://liblas.org/ libLAS] - LAS 1.0/1.1 ASPRS LiDAR data translation toolset | * [http://liblas.org/ libLAS] - LAS 1.0/1.1 ASPRS LiDAR data translation toolset | ||
* [http://pdal.io/ PDAL] - Point Data Abstraction Library | |||
* [http://code.google.com/p/fullanalyze/ Fullanalyze] software based on MATIS | |||
== Related publications == | |||
* Petras, V., Petrasova, A., Jeziorska, J., & Mitasova, H., 2016. Processing UAV and lidar point clouds in GRASS GIS. ISPRS-International Archives of the Photogrammetry, Remote Sensing and Spatial Information Sciences, 945-952. (PDF: [http://www.int-arch-photogramm-remote-sens-spatial-inf-sci.net/XLI-B7/945/2016/ ISPRS Archives], [https://www.researchgate.net/publication/304340172_Processing_UAV_and_lidar_point_clouds_in_GRASS_GIS ResearchGate]) | |||
* Brovelli, M.A., Lucca, S., 2012. Comparison of GRASS-LiDAR modules–TerraScan with respect to vegetation filtering. Appl Geomat 4, 123–134. ([http://link.springer.com/article/10.1007%2Fs12518-012-0080-6 PDF]) | |||
* Brovelli, M.A., Lucca, S., 2011. Filtering LiDAR with GRASS: overview of the method and comparisons with Terrascan. Italian Journal of Remote Sensing 93-105. ([http://www.aitjournal.com/articleView.aspx?ID=209 PDF]) | |||
* Brovelli, M.A., Cannata, M. & Longoni, U.M., 2004. LIDAR data filtering and DTM interpolation within GRASS. Transactions in GIS, 8(2), pp.155-174. [http://onlinelibrary.wiley.com/doi/10.1111/j.1467-9671.2004.00173.x/pdf PDF] | |||
* Cebecauer, T., Hofierka, J. & Suri, M., 2002. Processing digital terrain models by regularized spline with tension: tuning interpolation parameters for different input datasets. In Proc. of the Open Source Free Software GIS -- GRASS users conference 2002, Trento, Italy, 11-13 September 2002. [http://www.ing.unitn.it/~grass/conferences/GRASS2002/proceedings/proceedings/pdfs/Cebecauer_Tomas.pdf PDF] | |||
* Mitasova, Helena et al., 2009. Raster-based analysis of coastal terrain dynamics from multitemporal Lidar data. Journal of Coastal Research, 25(2), pp.507-514. | |||
* Mitasova, H, Mitas, L. & Harmon, R., 2005. Simultaneous spline approximation and topographic analysis for lidar elevation data in open-source GIS. Geoscience and Remote Sensing Letters, IEEE, 2(4), pp.379, 375. [http://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.69.2934&rep=rep1&type=pdf PDF] | |||
* Mitasova, H. et al., 2003. Spatio-temporal analysis of beach morphology using LIDAR, RTK-GPS and Open source GRASS GIS. In Proceedings Coastal Sediments. [http://skagit.meas.ncsu.edu/%7Ehelena/publwork/papers/pcoastsedp10.pdf PDF] | |||
Search via [http://scholar.google.com/scholar?q=lidar+grass+gis Google Scholar] | |||
== See also == | |||
* [[Lidar Analysis of Vegetation Structure]] | |||
* [[Processing lidar and UAV point clouds in GRASS GIS (workshop at FOSS4G Boston 2017)]] | |||
* [[Introduction to GRASS GIS with terrain analysis examples]] | |||
* [[From GRASS GIS novice to power user (workshop at FOSS4G Boston 2017)]] | |||
* [http://web.archive.org/web/20120324184538/http://knol.google.com/k/aerial-extraction-of-roof-surfaces-for-solar-analysis Aerial Extraction of Roof Surfaces for Solar Analysis] | |||
[[Category:Documentation]] | [[Category:Documentation]] | ||
[[Category: Interpolation]] | |||
[[Category: Image processing]] | |||
[[Category: Import]] | |||
[[Category: Raster]] | |||
[[Category: Vector]] | |||
Latest revision as of 22:50, 11 October 2020

LIDAR and Multi-beam Swath bathymetry data
Point cloud data, as a type of representation of 3D surfaces, are usually produced by airborne or on-ground laser scanning, also known as Light Detection and Ranging (LiDAR). The data are often provided as sets of very dense (x, y, z) points or in a more complex, public file binary format called LAS that may include multiple returns as well as intensities. GRASS GIS supports basic and advanced lidar data processing and analysis.
Modules
In this section various modules are introduced.
Import
- r.in.xyz - Create a raster map from an assemblage of many coordinates using univariate statistics. (example)
- r.in.lidar - (GRASS 7 only; GRASS must be compiled with libLAS support) Create a raster map from a binary LAS format LiDAR file (*.las) using univariate statistics and filtering. r.in.lidar is based on r.in.xyz. In addition to the options of r.in.xyz, r.in.lidar provides some basic lidar point filter options.
- v.in.ascii - Import data from an ASCII file to GRASS vector format.
- Due to memory overhead vector point imports will be limited to a few million data points unless topology and database creation is skipped with the -bt flags. It may also be useful to clip the import file to only accept points falling within the current region by using the -r flag. See g.region for details on specifying the region bounds.
- v.in.lidar - (GRASS 7 only; GRASS must be compiled with libLAS support). Creates a vector points file from a binary LAS format LiDAR file (*.las or *.laz). r.in.lidar also can create a new location based on the LAS file, and can filter the input points by return and subregion.
Analysis
- v.outlier - Removes outliers from vector point data.
- v.lidar.edgedetection - Uses interpolation and edge detection to create a new vector points file of LiDAR data so that the resulting attribute table is reclassified with CAT=1 for points associated with the ground surface (i.e., terrain) and useful for interpolating a raster terrain (DEM) map, CAT=2 for points pertaining to edges of human-contructed objects, and CAT=3 for other points that could pertain to vegetation or other features.
- v.lidar.growing - Building contour determination and region growing algorithm for determining the building inside.
- v.lidar.correction - Correction of the v.lidar.growing output. It is the last of the three algorithms for LIDAR filtering.
References:
- Detailed description: here (FOSS4G 2006)
- Summarised version: LiDAR filtering with GRASS (2007)
- Calibration of the filtering parameters (around 20 parameters) by integrating the USGS UCODE and GRASS, see INVERSE CALIBRATION OF LIDAR FILTERING PARAMETERS: UCODE/GRASS INTEGRATION
- See also the Related publications section below
Surface generation
- v.surf.rst - Spatial approximation and topographic analysis using regularized spline with tension.
- v.surf.idw - Surface interpolation from vector point data by Inverse Distance Squared Weighting.
- v.surf.bspline - Surface interpolation from vector point data by bicubic or bilineal interpolation with Tykhonov regularization.
- r.fillnulls - Fills no-data areas in raster maps using v.surf.rst splines interpolation.
- r.surf.nnbathy - Natural Neighbor interpolation using the 'nn' addon.
LAS support
The commands r.in.lidar and v.in.lidar offer LAS support. This requires that GRASS GIS is compiled with liblas (optionally laszip).
# installation on Fedora/Centos/EPEL7 yum install liblas liblas-devel laszip # installation on Ubuntu apt-get install liblas-bin liblas-dev
Configuration of GRASS GIS for libLAS support:
./configure \ ... --with-liblas ...
Then Compile and Install.
Swath Bathymetry Tools
see also the Marine_Science#Multibeam_sonar_processing wiki page
- The v.swathwidth module by David Finlayson for planning surveys. (development page)
- An example of post-processing scanned paper sidescan swaths using thin plate spline warping with GDAL's "gdalwarp -tps" function. (debugging page)
- GRASS integration with MB-System (GPL) software for processing Multibeam and Sidescan Sonar data. GRASS + MBsys + GMT make a nice scriptable trio.
LIDAR Tools
- r.terraflow - computation of flow direction, flow accumulation and other basic topographic terrain indices from massive raster digital elevation models (DEM). From the Duke University STREAM project.
- Flood simulation using r.lake. Includes fancy NVIZ visualization of Trento, Italy, by Markus Neteler.
- LAStools are a set of simple command line tools for converting to/from ASCII, viewing, comparing, and compressing LIDAR data. While free to use source code is available for older verions, newer versions are not open source, only work on MS Windows, and are no longer free for commercial or government use.
- libLAS ASPRS LiDAR data translation tools
- See also LIDAR Analysis
Micro-tutorial for LAS data import
The following scripts are given for UNIX Bourne Shell; MS-Windows users should use the Msys terminal to use them.
Preparation
Conversion of text files to LAS
(optional: While you do not need to do this conversion for GRASS import, the resulting files are much smaller than the uncompressed text files; additionally, they are in a defined format. On the other hand ASCII text files will survive decades without the need for external software.)
libLAS supports the following column types:
x - x coordinate y - y coordinate z - z coordinate a - scan angle i - intensity n - number of returns for given pulse (1..n) r - number of this return (1..r) c - classification u - user data (but only 1 byte) p - point source ID e - edge of flight line d - direction of scan flag t - GPS time s - skip column
Sample text data such as:
returntime,pulse,east,north,height,intensity,stripe 549778.907200,1,673999.940,5099680.080,507.425,20.0,45105 ...
can be converted to LAS format like this:
returntime - t pulse - r east - x north - y height - z intensity - i stripe - s
First parse (sanity check):
txt2las -parse trxyzis data.asc
Then convert:
txt2las -parse trxyzis -i data.asc -o data.las
Import
- For GRASS 6 use the libLAS utilities to convert LAS data into ASCII text format for GRASS.
- Data stored in text files (for example, .csv) can generally be imported directly into GRASS.
- The data used in the rest of this micro-tutorial can be found at appliedimagery.com.
Import ASCII text into a raster DEM
Check bounds of LAS file
Check bounds and SRS:
$ lasinfo "Serpent_Mound_Model_LAS_Data.las" [...] Min X Y Z 289020.900000 4320942.610000 166.780000 Max X Y Z 290106.020000 4323641.570000 215.480000 Spatial Reference +proj=utm +zone=17 +ellps=WGS84 +units=m
Check bounds of ASCII text file
This is similar in form to the above, but use the '-s' scan flag with the r.in.xyz module. Add the '-g' flag to get output ready for g.region.
Set region bounds and grid size
After creating a suitable UTM zone 17 location (EPSG:32617; the Serpent_Mound is in Ohio, USA) set the region according to the information from lasinfo at 1m resolution, using the '-a' flag to round the grid outwards, aligning to whole meters:
GRASS> g.region n=4323641.57 s=4320942.61 w=289020.90 e=290106.02 res=1 -ap
Run the import
Finally, import with r.in.xyz with data piped directly from the las2txt program and set a nice equalized color table:
BASEMAP="Serpent_Mound_Model_LAS"
las2txt --stdout "${BASEMAP}_Data.las" | \
r.in.xyz in=- out=${BASEMAP}_Data fs=comma method=mean
r.colors ${BASEMAP}_Data color=bcyr -e
- The above example uses the default z-elevation level as the 3rd term, but by using the las2txt --parse command other fields (such as intensity) can be imported instead via "--parse xyi". Unfortunately there seem to be a number of versions of las2txt and each are called slightly differently. You might have to experiment a little to get the right incantation.
- To import from an ASCII text file, run r.in.xyz directly with input= set to the filename instead of "-" (which indicates input will be piped in from another program).
Direct import of LAS as raster DEM
This is the same as the above, but use the r.in.lidar module (only in GRASS 7). In all cases you should manually decide on a suitable grid resolution and bounds, then set them with g.region, before running the import modules.
Import LAS as vector points in GRASS 6
Region setting (establishing the grid) is not needed for vector features so we can go directly to the import step. To deal with millions of input points v.in.ascii should be run with the options to skip creation of an attribute database and building topology as these can consume large amounts of memory. Note that vector maps without topology built are somewhat limited in their ability to be processed. Most LIDAR specific modules have been adapted to not require topology. Even so, after initial cleaning steps it is often more efficient to work with huge datasets in GRASS as raster data.
las2txt --stdout "${BASEMAP}_Data.las" | \
v.in.ascii -tbz z=3 out="${BASEMAP}_pts" fs=comma
If topology was built, you can use d.vect's -z flag to colorize by elevation value. Without topology you can still colorize, but you need to use color rules based on absolute elevations, not percentage of scale.
# display colorized points for data with built topology d.vect map=lidar_pts size=1 -z zcolor=elevation
- After import as points you can then use the v.lidar tools to clean the data: v.lidar.correction, v.lidar.edgedetection, v.lidar.growing, and v.outlier.
- Vector points can be interpolated into raster DEMs with a number of modules, including v.surf.rst, v.surf.bspline, and v.surf.idw.
Import LAS in GRASS 7
As vector points
- GRASS 7 offers the v.in.lidar command
As raster: map:
- r.in.lidar command
- r.in.pdal (src)
Clean imported raster DEM
(fill holes)
# convert to vector points
r.to.vect -z feature=point in=${BASEMAP}_Data out=${BASEMAP}_pt
# interpolate using a regularized spline fit
# this is very slow, but produces very high quality output
v.surf.rst layer=0 in=${BASEMAP}_pt elev=${BASEMAP}.rst
# create 5m buffer area around original data points
r.buffer in=${BASEMAP}_Data out=${BASEMAP}.5m_buff dist=5
# crop interpolated DEM to only include areas nearby actual data
r.mapcalc "${BASEMAP}.filled = \
if( isnull(${BASEMAP}.5m_buff), null(), ${BASEMAP}.rst)"
# set colors to something nice
r.colors ${BASEMAP}.filled color=bcyr -e
Depending on your needs, r.fillnulls, v.surf.bspline, v.surf.idw, r.surf.idw, r.surf.idw2, or r.surf.nnbathy may be faster than the v.surf.rst method.
Visualize raster DEM in 3D
nviz ${BASEMAP}.filled
- Set z-exag to 2.0
- In Visualize → Raster Surfaces set the fine (final) resolution to 1, and coarse (preview) resolution to 5.
- Set the height to 500.0, the perspective to 15.0, and drag the view-puck to the North-West and reasonably zoomed in.
- You should now be able to see the serpent:
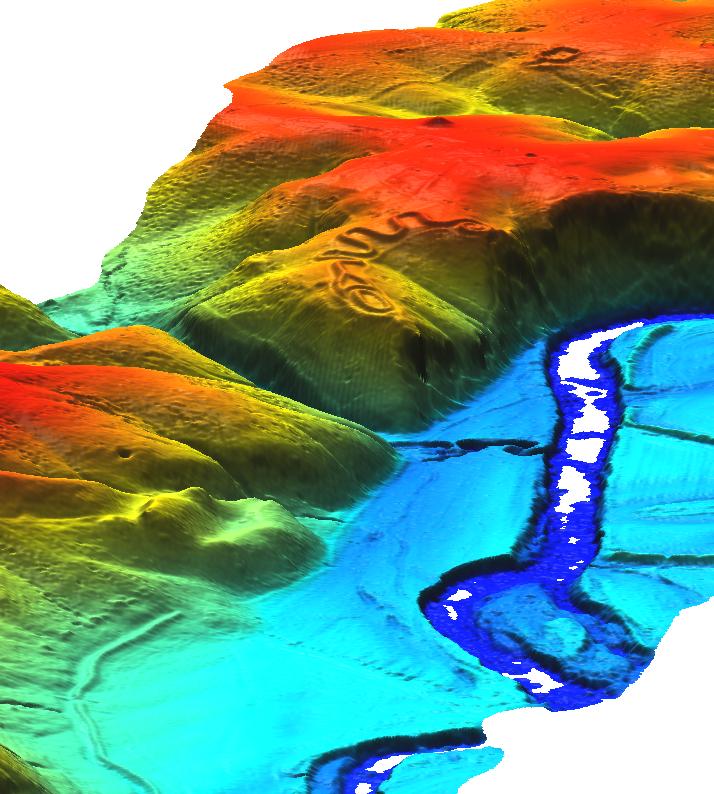
It is possible to show vector points in 3D, but millions of them may make the program run slow. Topology is required (v.build). Tick the "3D" box in the Visualize → Vector points dialog.
LAS Export
Export to LAS
This is the reverse of the import step, but using v.out.ascii or r.out.xyz with txt2las. Because the v.out.ascii module exports category number which we are not interested in, we cut it away with the UNIX cut utility.
v.out.ascii ${BASEMAP}_pts fs=space | cut -f1-3 -d' ' \
> ${BASEMAP}_export.txt
txt2las --parse xyz -i ${BASEMAP}_export.txt
Micro-tutorial for LIDAR data analysis
Simple analysis of single return data
Import from ASCII file
Data source: North Carolina sample data set (Lidar data from 2001, near Raleigh, North Carolina). Download from
http://www.grassbook.org/ncexternal/ -> File: BE3720079200WC20020829m.txt 3.6M (lidar bare earth points [spm])
Scan input file for spatial extent. The -g flag shows the result in a convenient copy-paste format for g.region:
r.in.xyz BE3720079200WC20020829m.txt out=dummy -s -g
We use this output to set region, predefine 2m raster cells, and polish the odd coordinates with -a:
g.region n=222504.439000 s=219456.442000 e=640081.274000 w=637033.274000 res=2 -a -p
QUESTION 1: Are these Lidar data sufficiently dense?
We compute a raster map representing number of points per cell
r.in.xyz BE3720079200WC20020829m.txt out=lid_792_binn2m method=n
Look at the resulting raster map:
d.mon x0 d.font Vera d.rast.leg lid_792_binn2m
Report point distribution in percent:
r.report lid_792_binn2m unit=p r.univar lid_792_binn2m # (consider running `r.null setnull=0` first)
Reduce the resolution to 6m to get at least one point per cell:
g.region res=6 -a -p r.in.xyz BE3720079200WC20020829m.txt out=lid_792_binn6m method=n d.rast.leg lid_792_binn6m # ... a few holes remain but that's probably acceptable.
Compute raster maps representing mean elevation for each cell:
r.in.xyz BE3720079200WC20020829m.txt out=lid_792_binmean6m \
method=mean
d.rast.leg lid_792_binmean6m
Compute range and variation:
r.in.xyz BE3720079200WC20020829m.txt out=lid_792_binrange6m \
method=range
d.rast.leg lid_792_binrange6m
r.in.xyz BE3720079200WC20020829m.txt out=lid_792_binvar6m \
method=coeff_var
d.rast.leg lid_792_binvar6m
Overlay other GIS maps to map of mean elevation:
d.rast.leg lid_792_binmean6m d.vect streets_wake d.vect lakes type=boundary d.vect streams
Now we continue to work in a zoomed spatial subset of the area.
Here we only import only points in the rural area, do not build topology, and use the z-coordinate for elevation:
g.region rural_1m -p
v.in.ascii -ztbr BE3720079200WC20020829m.txt \
out=elev_lidrural_bepts z=3
Clear the monitor, then show a black and white orthophoto:
d.erase -f d.rast ortho_2001_t792_1m
We now use two LiDAR vector point maps, which were prepared earlier:
- elev_lid792_bepts: Rural area (in tile 792), bare earth lidar point cloud
- elev_lidrural_mrpts: Rural area multiple return lidar point cloud
Look at ground data:
d.vect elev_lidrural_bepts size=2 color=red
Look at "surface" data:
d.vect elev_lidrural_mrpts size=1 color=green
Visualize 3D LiDAR multi-return points:
nviz elev_lid792_1m point=elev_lidrural_mrpts
# Visualize -> Vector Points -> 3D points # -> Icon size 2.25 # --> GRASS book p. 253, fig. 3.15 (see also screenshot here)
DEM/DSM separation the simple way by selection of Lidar returns
Background on lidar multiple returns: "Lidar returns are discrete observations* recorded when a laser pulse is intercepted and reflected by targets. Multiple returns derive from one laser pulse intercepting multiple targets (e.g. a top of a tree, its branches, and the ground)." (source: stackexchange).
Find out where we have multiple returns:
d.rast ortho_2001_t792_1m d.vect elev_lidrural_mrpts where="return=1" color=red size=2 d.vect elev_lidrural_mrpts where="return=2" color=green size=3 d.vect elev_lidrural_mrpts where="return=3" color=blue d.vect elev_lidrural_mrpts where="return=4" color=yellow
DTM: extract last return(s):
v.extract elev_lidrural_mrpts out=elev_lidrural_mrpts_first where="Return < 2" nviz elev_lid792_1m point=elev_lidrural_mrpts_first
Interpolate to map and look at it:
v.surf.rst elev_lidrural_mrpts_first layer=0 elev=elev_lidrural_mrpts_DTM nviz elev_lidrural_mrpts_DTM col=ortho_2001_t792_1m
DSM: extract first return(s):
v.extract elev_lidrural_mrpts out=elev_lidrural_mrpts_last where="Return > 2" nviz elev_lid792_1m point=elev_lidrural_mrpts_last
Interpolate to map and look at it:
v.surf.rst elev_lidrural_mrpts_first layer=0 elev=elev_lidrural_mrpts_DSM nviz elev_lidrural_mrpts_DSM col=ortho_2001_t792_1m
DEM/DSM separation the more complex way
TODO: verify order of first and last returns in below text
General procedure:
- Lidar point clouds (first and last return) are imported with v.in.ascii (-b flag to not build the topology).
- Outlier detection is done with v.outlier on both first and last return data (NB currently a selection of </=4 for soe/son in v.outlier results in an error message).
- Then, with v.lidar.edgedetection, edges are detected from last return data.
- The building are generated by v.lidar.growing from detected edges.
- The resulting data are post-processed with v.lidar.correction.
- Finally, the DTM and DSM are generated with v.surf.bspline (DTM: uses the 'v.lidar.correction' output; DSM: uses last return output from outlier detection).
- NB for v.outlier, v.lidar.edgedetection and v.surf.bspline, one spline steps equates to 1m. It is recommended as a starting point that the choice of spline step is roughly 3 or 4 times the planimetric resolution (potential grid resolution) of your data. Experiment from there to obtain better results.
export GRASS_OVERWRITE=1
v.extract elev_lidrural_mrpts out=elev_lidfirst_pts \
where="return = 1"
v.extract elev_lidrural_mrpts out=elev_lidlast_pts \
where="return >= 2"
d.vect elev_lidfirst_pts col=red
d.vect elev_lidlast_pts col=green
Outlier detection and separation into two maps
# 1st return
v.outlier elev_lidfirst_pts output=elev_lidfirst_clean \
outlier=elev_lidfirst_outl
d.erase
d.vect elev_lidfirst_clean size=2
d.vect elev_lidfirst_outl color=red
2nd return
v.outlier elev_lidlast_pts output=elev_lidlast_clean \
outlier=elev_lidlast_outl
d.erase
d.vect elev_lidlast_clean size=2
d.vect elev_lidlast_outl color=red
# -> no outliers visible
Run an edge detection on cleaned last return:
v.lidar.edgedetection elev_lidlast_clean \
out=elev_lidlast_edges
Buildings/vegetation are generated from detected edges (bug: you may need to specify the mapset):
v.lidar.growing elev_lidlast_edges@lidar out=elev_lidlast_grow \
first=elev_lidfirst_clean
Compare:
d.vect elev_lidlast_pts col=blue d.vect elev_lidlast_grow col=green
Correction (this is applied twice):
v.lidar.correction elev_lidlast_grow out=elev_lidlast_corr1 \
terrain=elev_lidlast_terr1
v.lidar.correction elev_lidlast_corr1 out=elev_lid_dsm \
terrain=elev_lid_dtm
DEM and DSM are generated:
# Estimation of lambda_i parameter with cross validation (watch for RMS!)
# and note use of bicubic for DSM and bilinear for DTM here and below
v.surf.bspline -c elev_lid_dsm sie=100 sin=100 method=bicubic
v.surf.bspline -c elev_lid_dtm sie=100 sin=100 method=bilinear
# From the cross-validation, we select lambda with minimal RMS error:
# generate raster surfaces at 1m resolution
v.surf.bspline elev_lid_dsm raster=lidar_dsm lambda=0.1 method=bicubic
v.surf.bspline elev_lid_dtm raster=lidar_dtm lambda=0.0001 method=bilinear
d.rast lidar_dsm
d.rast lidar_dtm
nviz lidar_dsm,lidar_dtm \
col=ortho_2001_t792_1m,ortho_2001_t792_1m
...with the position slider you can visually separate DSM and DEM, increase z slider (Visualize -> Raster Surface -> Position).
Reclassification
As vector points
- v.reclass command
As raster map:
- r.reclass command
Sample LIDAR data
Widely used in GRASS tutorials
- Jockey's Ridge, NC, LIDAR dataset
- North Carolina OSGeo Edu data set (includes multi-return LIDAR data)
Other
- United States Antarctic Resource Center: LIDAR High-resolution DEM Final DATA Downloads
http://usarc.usgs.gov/lidar_dload.shtml
- LIDAR ALSM Research, Arizona State University Ative Tectonics, Research Group
http://lidar.asu.edu/research.html and http://www.geongrid.org/science/lidar.html
- USGS Center for LIDAR Information Coordination and Knowledge (aka CLICK) - USGS LiDAR point cloud distribution site
http://lidar.cr.usgs.gov
- Washington State Geospatial Data Archive, Mount Saint Helens - Lidar Data
https://wagda.lib.washington.edu/data/type/elevation/lidar/st_helens/
- Puget Sound Lidar Consortium, public-domain high-resolution topography for western Washington
http://pugetsoundlidar.ess.washington.edu/index.htm
- NOAA Topographic Change Mapping LIDAR Data Retrieval Tool (LDART) NOAA Coastal Services Center
http://maps.csc.noaa.gov/TCM/
- Landmap, LIDAR Data from the Environment Agency
http://www.landmap.ac.uk/lidar/lidar.html
- Northern California LIDAR data
http://quake.usgs.gov/research/geology/lidar/ and http://core2.gsfc.nasa.gov/lidar/terrapoint/
- IDAHO GEOSPATIAL , Bare Earth LIDAR DEM Download - UTM
http://inside.uidaho.edu/geodata/LiDAR/LiDARBareEarthDEM_DownloadUTM.htm
- EarthScope Spatial Data Explorer - A java application for querying, browsing, and acquiring data from the EarthScope Spatial Data Repository. Currently includes a number of LiDAR datasets.
http://www.earthscope.org/data/lidar.php
- South Tyrol - Download of DTMs (Homepage in German or Italian)
http://www.provinz.bz.it/raumordnung/grundkarten/utm/default_d.htm
- libLAS's sample file collection
- NASA's Laser Vegetation Imaging Sensor (a.k.a. the Land, Vegetation, and Ice Sensor) or "LVIS"
Tutorials
- Lidar Analysis of Vegetation Structure
- Processing lidar and UAV point clouds in GRASS GIS
- Lectures and assignments from NCSU partially dealing with lidar
- Some basic and some new (7.2-7.4) GRASS GIS features related to lidar
- Breaklines
- Not very well developed in GRASS so far, but we'd like to change that (you can volunteer)
- v.surf.icw (only suitable for ~ 200 input points)
- SurfIt (GPL, Tck/Tk)
SurGe (Trial/shareware)- v.triangle add-on module for construction TIN with breaklines.
- see above Contour lines to DEM page for further discussion of the (in)appropriateness of using TINs to generate raster surfaces. It is hoped that in future GRASS's more advanced spline interpolation modules will get breakline support.
Links
- Contour lines to DEM interpolation module trials
- libLAS - LAS 1.0/1.1 ASPRS LiDAR data translation toolset
- PDAL - Point Data Abstraction Library
- Fullanalyze software based on MATIS
Related publications
- Petras, V., Petrasova, A., Jeziorska, J., & Mitasova, H., 2016. Processing UAV and lidar point clouds in GRASS GIS. ISPRS-International Archives of the Photogrammetry, Remote Sensing and Spatial Information Sciences, 945-952. (PDF: ISPRS Archives, ResearchGate)
- Brovelli, M.A., Lucca, S., 2012. Comparison of GRASS-LiDAR modules–TerraScan with respect to vegetation filtering. Appl Geomat 4, 123–134. (PDF)
- Brovelli, M.A., Lucca, S., 2011. Filtering LiDAR with GRASS: overview of the method and comparisons with Terrascan. Italian Journal of Remote Sensing 93-105. (PDF)
- Brovelli, M.A., Cannata, M. & Longoni, U.M., 2004. LIDAR data filtering and DTM interpolation within GRASS. Transactions in GIS, 8(2), pp.155-174. PDF
- Cebecauer, T., Hofierka, J. & Suri, M., 2002. Processing digital terrain models by regularized spline with tension: tuning interpolation parameters for different input datasets. In Proc. of the Open Source Free Software GIS -- GRASS users conference 2002, Trento, Italy, 11-13 September 2002. PDF
- Mitasova, Helena et al., 2009. Raster-based analysis of coastal terrain dynamics from multitemporal Lidar data. Journal of Coastal Research, 25(2), pp.507-514.
- Mitasova, H, Mitas, L. & Harmon, R., 2005. Simultaneous spline approximation and topographic analysis for lidar elevation data in open-source GIS. Geoscience and Remote Sensing Letters, IEEE, 2(4), pp.379, 375. PDF
- Mitasova, H. et al., 2003. Spatio-temporal analysis of beach morphology using LIDAR, RTK-GPS and Open source GRASS GIS. In Proceedings Coastal Sediments. PDF
Search via Google Scholar
See also
- Lidar Analysis of Vegetation Structure
- Processing lidar and UAV point clouds in GRASS GIS (workshop at FOSS4G Boston 2017)
- Introduction to GRASS GIS with terrain analysis examples
- From GRASS GIS novice to power user (workshop at FOSS4G Boston 2017)
- Aerial Extraction of Roof Surfaces for Solar Analysis