Processing lidar and UAV point clouds in GRASS GIS (workshop at FOSS4G Boston 2017)

Description: GRASS GIS offers, besides other things, numerous analytical tools for point clouds, terrain, and remote sensing. In this hands-on workshop we will explore the tools in GRASS GIS for processing point clouds obtained by lidar or through processing of UAV imagery. We will start with a brief and focused introduction into GRASS GIS graphical user interface (GUI) and we will continue with short introduction to GRASS GIS Python interface. Participants will then decide if they will use GUI, command line, Python, or online Jupyter Notebook for the rest of the workshop. We will explore the properties of the point cloud, interpolate surfaces, and perform advanced terrain analyses to detect landforms and artifacts. We will go through several terrain 2D and 3D visualization techniques to get more information out of the data and finish with vegetation analysis.
Requirements: This workshop is accessible to beginners, but some basic knowledge of lidar processing or GIS is helpful for a smooth experience.
Authors: Vaclav Petras, Anna Petrasova, and Helena Mitasova from North Carolina State University
Contributors: Robert S. Dzur and Doug Newcomb
Preparation
Software
GRASS GIS 7.2 compiled with libLAS is needed (e.g. r.in.lidar should work).
OSGeo-Live
All needed software is included in OSGeo-Live.
Ubuntu
Install GRASS GIS from packages:
sudo add-apt-repository ppa:ubuntugis/ubuntugis-unstable sudo apt-get update sudo apt-get install grass grass-dev
Linux
For other Linux distributions other then Ubuntu, please try to find GRASS GIS in their package managers or refer to grass.osgeo.org.
MS Windows
Download the standalone GRASS GIS binaries from grass.osgeo.org.
Mac OS
Install GRASS GIS using Homebrew osgeo4mac:
brew tap osgeo/osgeo4mac brew install numpy brew install liblas --build-from-source brew install grass7 --with-liblas
If you have these already installed, you need to use reinstall instead of install.
Note that on Mac OS in some versions the r.in.lidar module is not accessible, so you need to check this and use r.in.ascii in combination with libLAS or PDAL command line tools to achieve the same or, preferably, use OSGeo-Live.
Note also that there is currently no recent version of installation on Mac OS where 3D view is accessible.
Addons
You will need to install the following GRASS GIS Addons. This can be done through GUI, but for simplicity copy and paste and execute the following lines one by one in the command line:
g.extension r.geomorphon g.extension r.skyview g.extension r.local.relief g.extension r.shaded.pca g.extension r.area g.extension r.terrain.texture g.extension r.fill.gaps
For bonus tasks:
g.extension v.lidar.mcc
Data
We will need the two following ZIP files, downloaded and extracted:
Later, for bonus task, download also this (much larger) file:
Basic introduction to graphical user interface
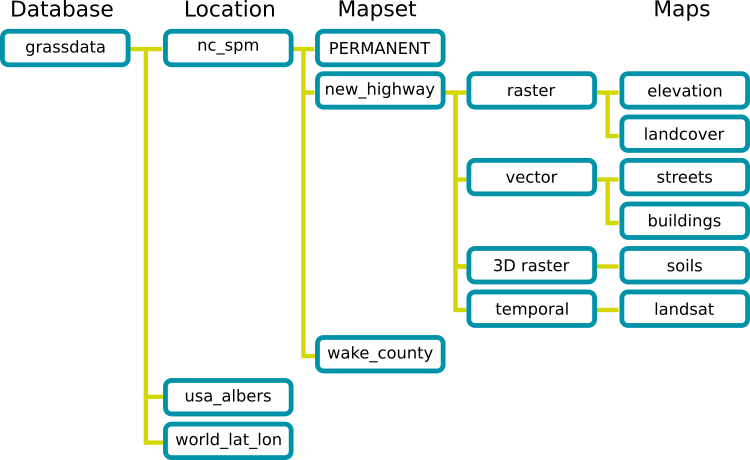
GRASS GIS Spatial Database
Here we provide an overview of GRASS GIS. For this exercise it's not necessary to have a full understanding of how to use GRASS GIS. However, you will need to know how to place your data in the correct GRASS GIS database directory, as well as some basic GRASS functionality.
GRASS uses specific database terminology and structure (GRASS GIS Spatial Database) that are important to understand for working in GRASS GIS efficiently. You will create a new location and import the required data into that location. In the following we review important terminology and give step by step directions on how to download and place your data in the correct place.
- A GRASS GIS Spatial Database (GRASS database) consists of directory with specific Locations (projects) where data (data layers/maps) are stored.
- Location is a directory with data related to one geographic location or a project. All data within one Location has the same coordinate reference system.
- Mapset is a collection of maps within Location, containing data related to a specific task, user or a smaller project.
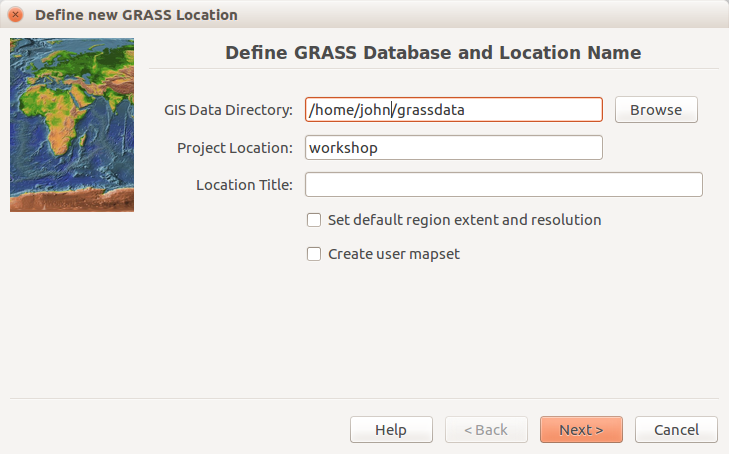
Start GRASS GIS, a start-up screen should appear. Unless you already have a directory called grassdata in your Documents directory (on MS Windows) or in your home directory (on Linux), create one. You can use the Browse button and the dialog in the GRASS GIS start up screen to do that.
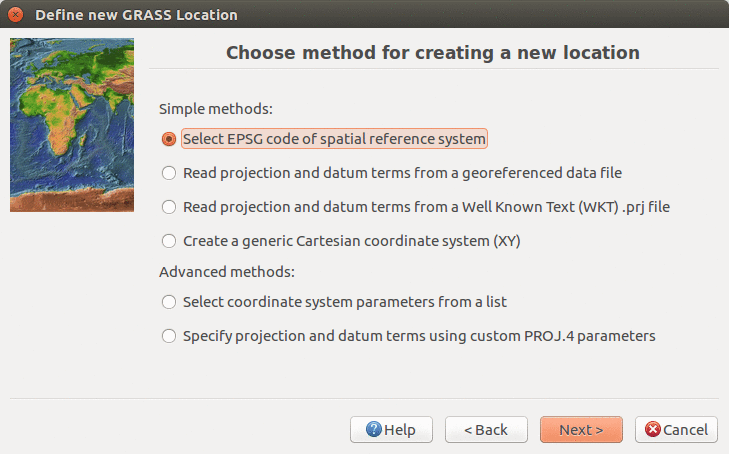
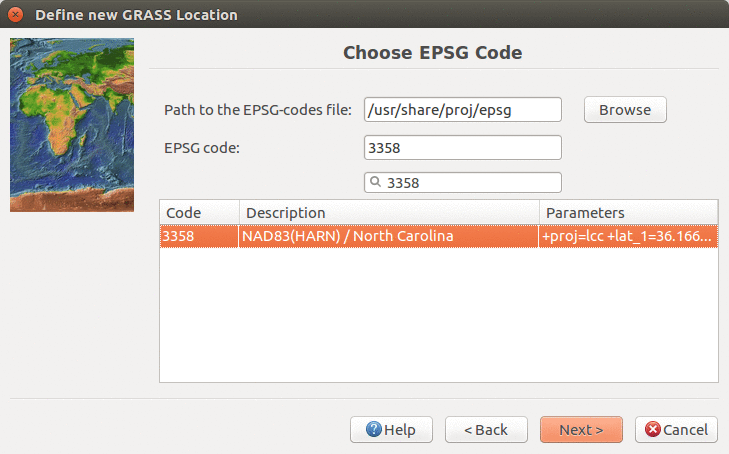
We will create a new location for our project with CRS (coordinate reference system) NC State Plane Meters with EPSG code 3358. Open Location Wizard with button New in the left part of the welcome screen. Select a name for the new location, select EPSG method and code 3358. When the wizard is finished, the new location will be listed on the start-up screen. Select the new location and mapset PERMANENT and press Start GRASS session.
-
GRASS GIS Spatial Database structure
-
GRASS GIS 7.2 startup dialog
-
Start Location Wizard and type the new location's name
-
Select method for describing CRS
-
Find and select EPSG 3358
-
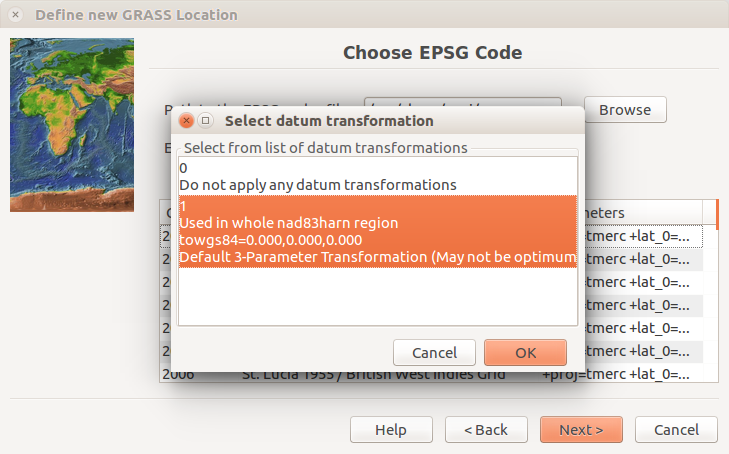
Confirm the use of the default datum transformation
-
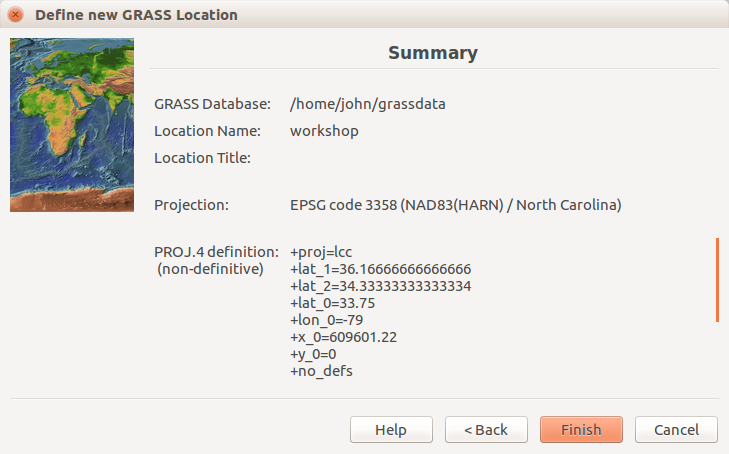
Review summary page and confirm
Note that a current working directory is a concept which is separate from the GRASS database, location and mapset discussed above. The current working directory is a directory where any program (no just GRASS GIS) writes and reads files unless a path to the file is provided. The current working directory can be changed from the GUI using Settings → GRASS working environment → Change working directory or from the Console using the cd command. This is advantageous when we are using command line and working with the same file again and again. This is often the case when working with lidar data. We can change the directory to the directory with the downloaded LAS file. IN case we don't change the directory, we need to provide full path to the file. Note that command line and GUI have their own settings of current working directory, so it needs to be changed separately for each of them.
Importing data
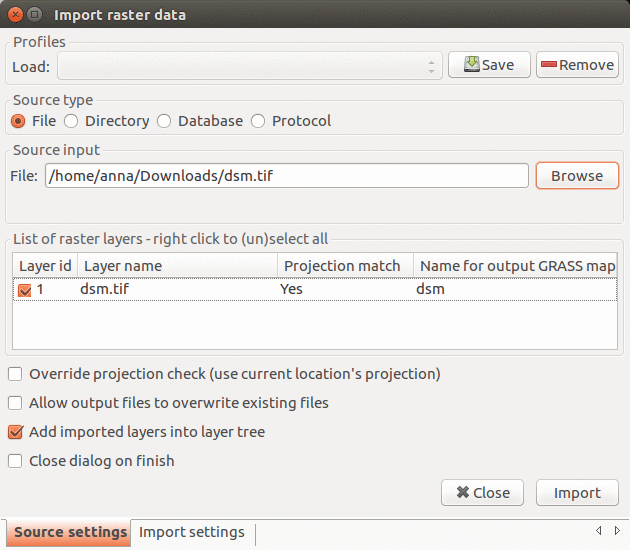
In this step we import the provided data into GRASS GIS. In menu File - Import raster data select Common formats import and in the dialog browse to find the orthophoto file, change the name to ortho, and click button Import. All the imported layers should be added to GUI automatically, if not, add them manually. Point clouds will be imported later in a different way as part of the analysis.
-
Import raster data: select the orthophoto file and change the name to ortho
The equivalent command is:
r.import input=nc_orthophoto_1m_spm.tif output=ortho
Computational region
Before we use a module to compute a new raster map, we must properly set the computational region. All raster computations will be performed in the specified extent and with the given resolution.
Computational region is an important raster concept in GRASS GIS. In GRASS a computational region can be set, subsetting larger extent data for quicker testing of analysis or analysis of specific regions based on administrative units. We provide a few points to keep in mind when using the computational region function:
- defined by region extent and raster resolution
- applies to all raster operations
- persists between GRASS sessions, can be different for different mapsets
- advantages: keeps your results consistent, avoid clipping, for computationally demanding tasks set region to smaller extent, check that your result is good and then set the computational region to the entire study area and rerun analysis
- run
g.region -por in menu Settings - Region - Display region to see current region settings
-
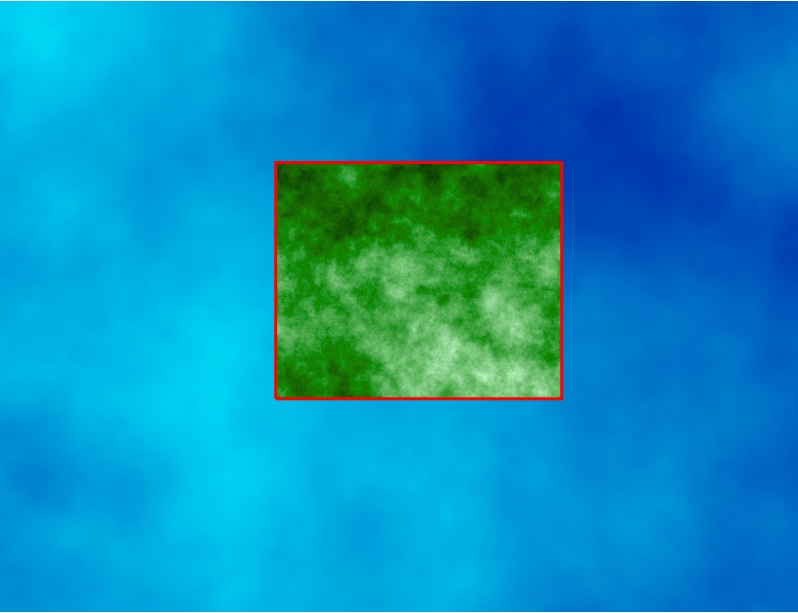
Computational region concept: A raster with large extent (blue) is displayed as well as another raster with smaller extent (green). The computational region (red) is now set to match the smaller raster, so all the computations are limited to the smaller raster extent even if the input is the larger raster. (Not shown on the image: Also the resolution, not only the extent, matches the resolution of the smaller raster.)
-
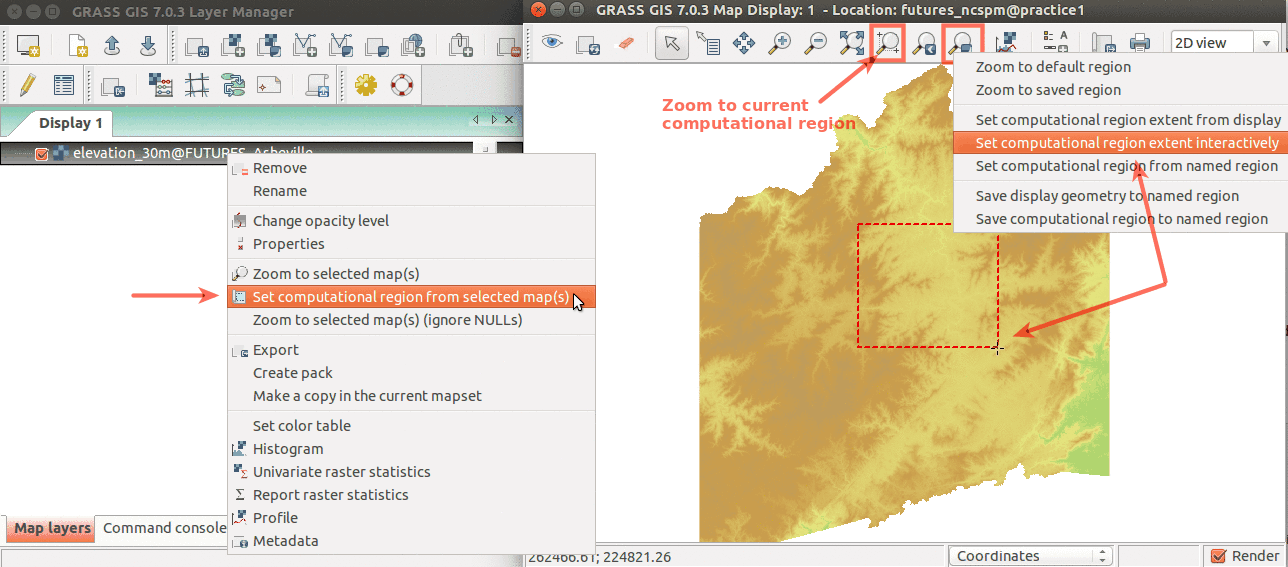
Simple ways to set computational region from GUI: On the left, set region to match raster map. On the right, select the highlighted option and then set region by drawing rectangle.
-
Set computational region (extent and resolution) to match a raster (Layers tab in the Layer Manager)
The numeric values of computational region can be checked using:
g.region -p
After executing the command you will get something like this:
north: 220750 south: 220000 west: 638300 east: 639000 nsres: 1 ewres: 1 rows: 750 cols: 700 cells: 525000
Computational region can be set using a raster map:
g.region raster=ortho -p
Resolution can be set separately using the res parameter of the g.region module. The units are the units of the current location, in our case meters. This can be done in the Resolution tab of the g.region dialog or in the command line in the following way (using also the -p flag to print the new values):
g.region res=3 -p
The new resolution may be slightly modified in this case to fit into the extent which we are not changing. However, often we want the resolution to be the exact value we provide and we are fine with a slight modification of the extent. That's what -a flag is for. On the other hand, if alignment with cells of a specific raster is important, align parameter can be used to request the alignment to that raster (regardless of the the extent).
The following example command will use the extent from the raster named ortho, use resolution 5 meters, modify the extent to align it to this 5 meter resolution, and print the values of this new computational region settings:
g.region raster=ortho res=5 -a -p
Modules
GRASS GIS functionality is available through modules (which are sometimes called tools, functions, or commands). Modules respect the following naming conventions:
| Prefix | Function | Example |
|---|---|---|
| r. | raster processing | r.mapcalc: map algebra |
| v. | vector processing | v.surf.rst: surface interpolation |
| i. | imagery processing | i.segment: image segmentation |
| r3. | 3D raster processing | r3.stats: 3D raster statistics |
| t. | temporal data processing | t.rast.aggregate: temporal aggregation |
| g. | general data management | g.remove: removes maps |
| d. | display | d.rast: display raster map |
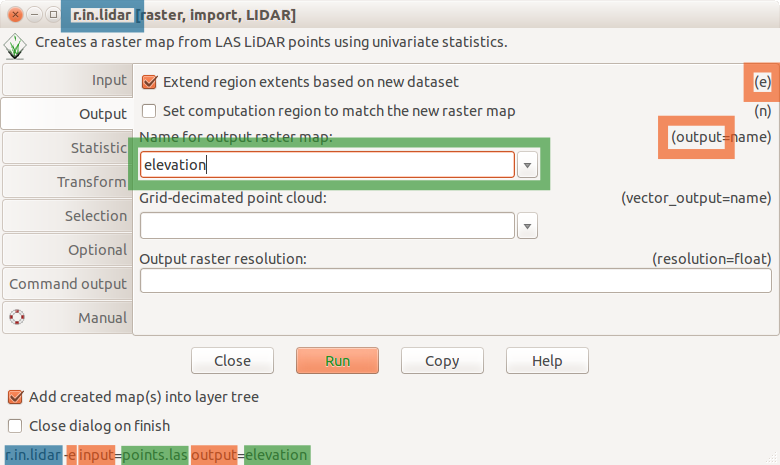
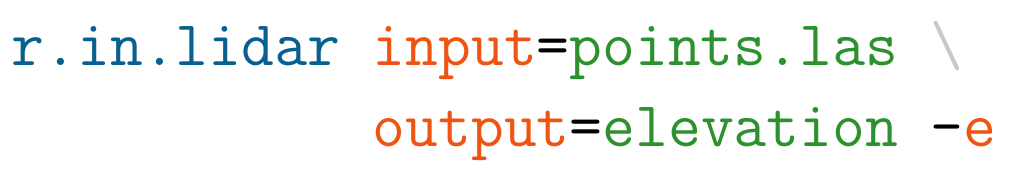
These are the main groups of modules. There is few more for specific purposes. Note also that some modules have multiple dots in their names. This often suggests further grouping. For example, modules staring with v.net. deal with vector network analysis. The name of the module helps to understand its function, for example v.in.lidar starts with v so it deals with vector maps, the name follows with in which indicates that the module is for importing the data into GRASS GIS Spatial Database and finally lidar indicates that it deals with lidar point clouds.
-
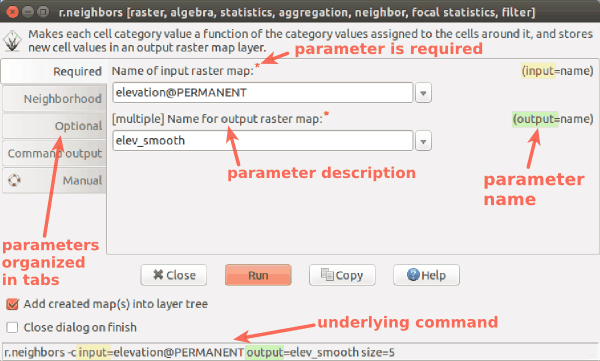
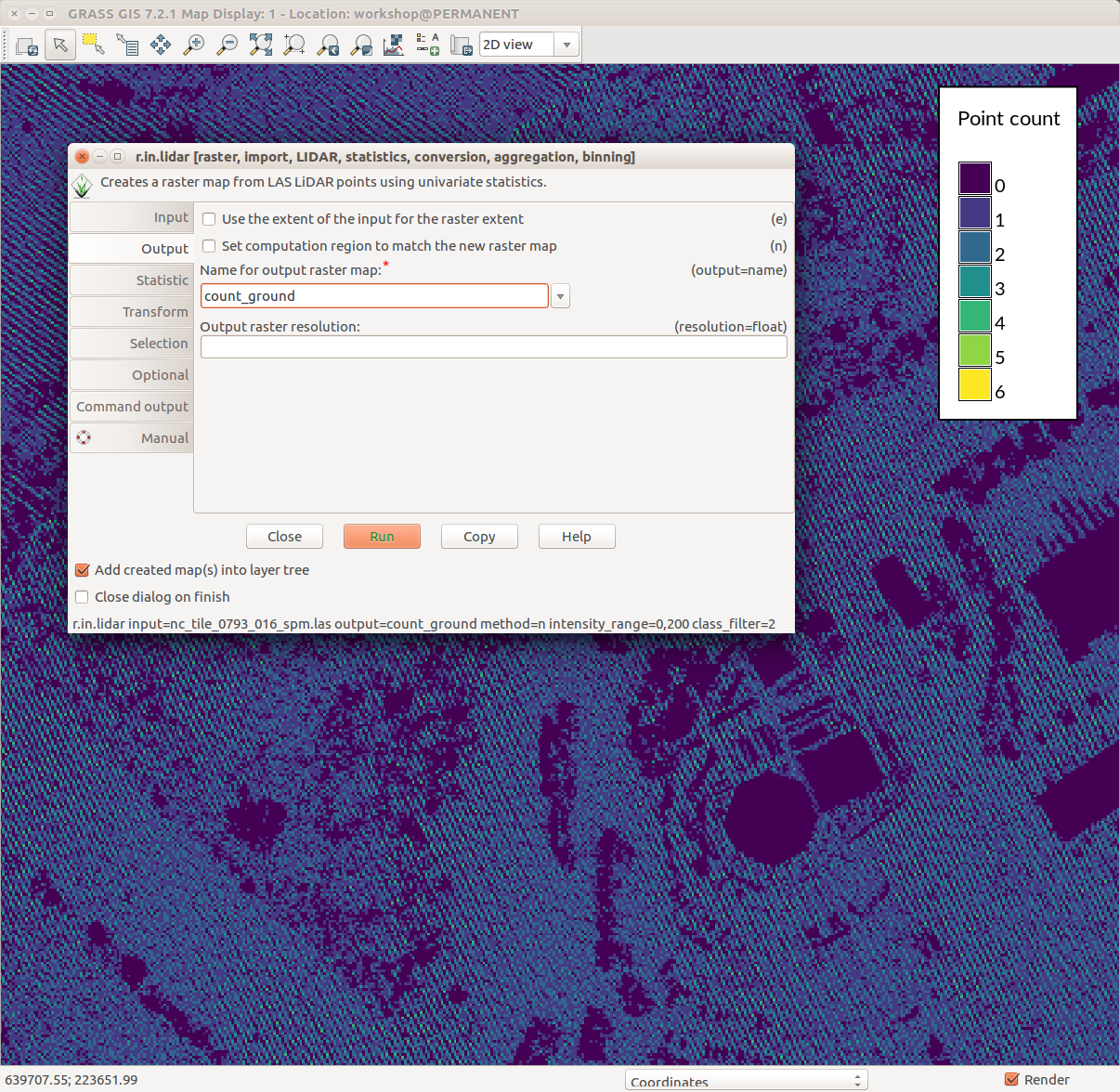
r.in.lidar dialog with Output tab active and highlighted module name (blue), options and flags (red) and option values (green)
-
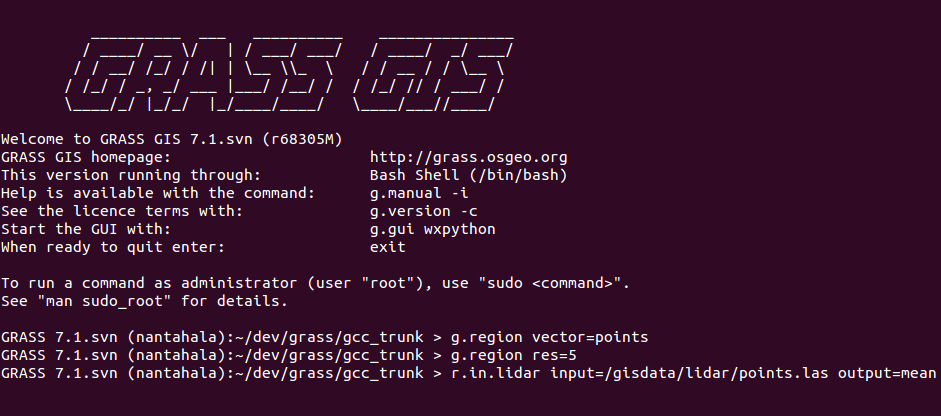
Example r.in.lidar command in Bash with highlighted module name (blue), options and flags (red) and option values (green)
-
Example r.in.lidar command in Python with highlighted module name (blue), options and flags (red) and option values (green) and import (grey)
-
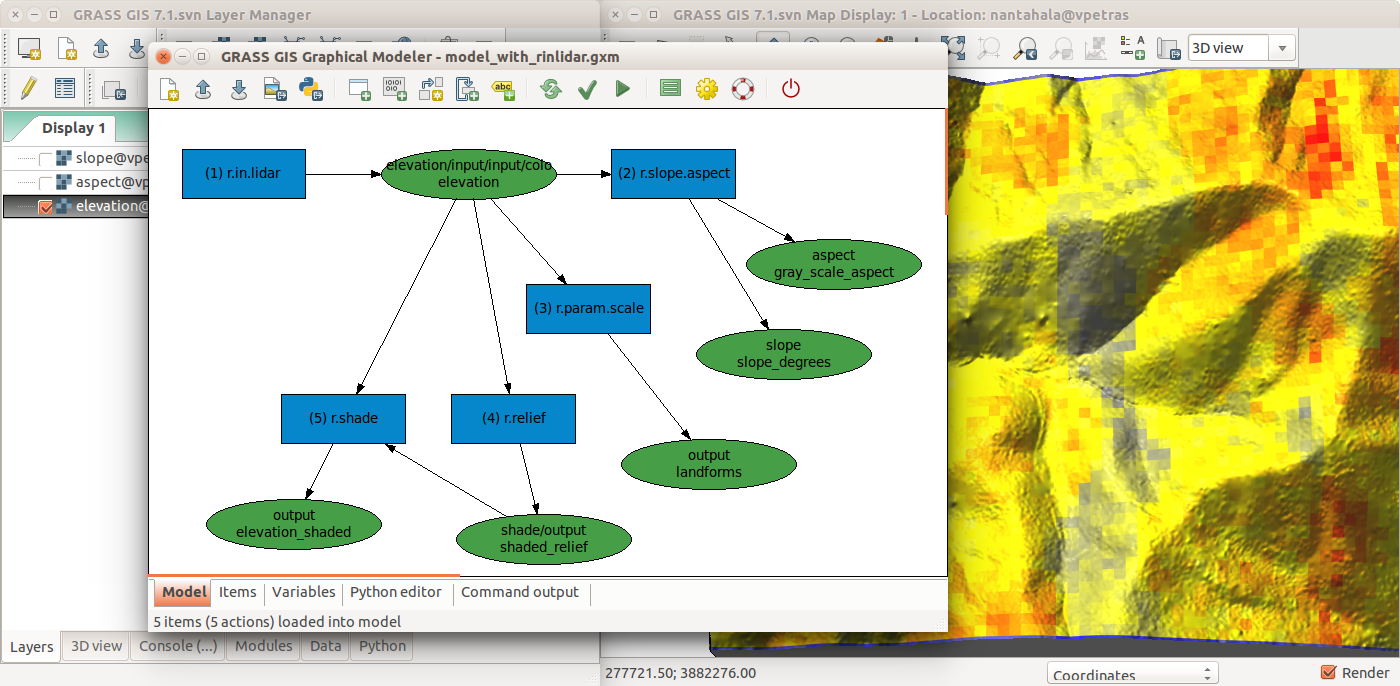
GRASS GIS Graphical Modeler
-
Console tab in the Layer Manager for running commands or opening module dialogs
-
Command line interface (CLI) in a system terminal
-
Layout of a module dialog
-
Search for a module in the Modules tab
-
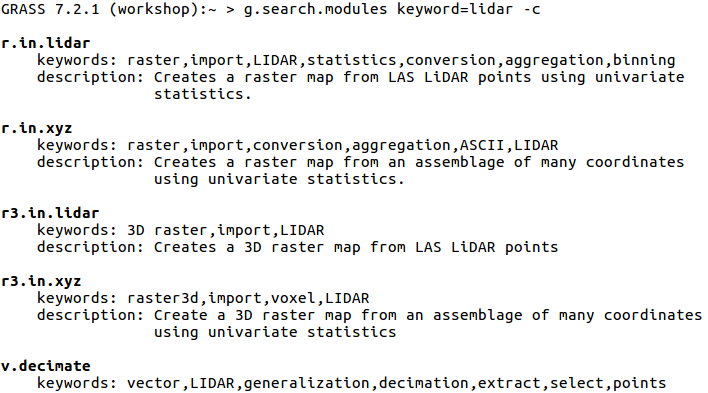
Search for a module using advanced search with g.search.modules
One of the advantages of GRASS GIS is the diversity and number of modules that let you analyze all manners of spatial and temporal data. GRASS GIS has over 500 different modules in the core distribution and over 300 addon modules that can be used to prepare and analyze data. The following table lists some of the main modules for point cloud analysis.
| Module | Function | Alternatives |
|---|---|---|
| r.in.lidar | binning into 2D raster, statistics | r.in.xyz, v.vect.stats, r.vect.stats |
| v.in.lidar | import, decimation | v.in.ascii, v.in.ogr, v.import |
| r3.in.lidar | binning into 3D raster | r3.in.xyz, r.in.lidar |
| v.out.lidar | export of point cloud | v.out.ascii, r.out.xyz |
| v.surf.rst | interpolation surfaces from points | v.surf.bspline, v.surf.idw |
| v.lidar.edgedetection | ground and object (edge) detection | v.lidar.mcc, v.outlier |
| v.decimate | decimate (thin) a point cloud | v.in.lidar, r.in.lidar |
| r.slope.aspect | topographic parameters | v.surf.rst, r.param.scale |
| r.relief | shaded relief computation | r.skyview, r.local.relief |
| r.colors | raster color table management | r.cpt2grass, r.colors.matplotlib |
| g.region | resolution and extent management | r.in.lidar, GUI |
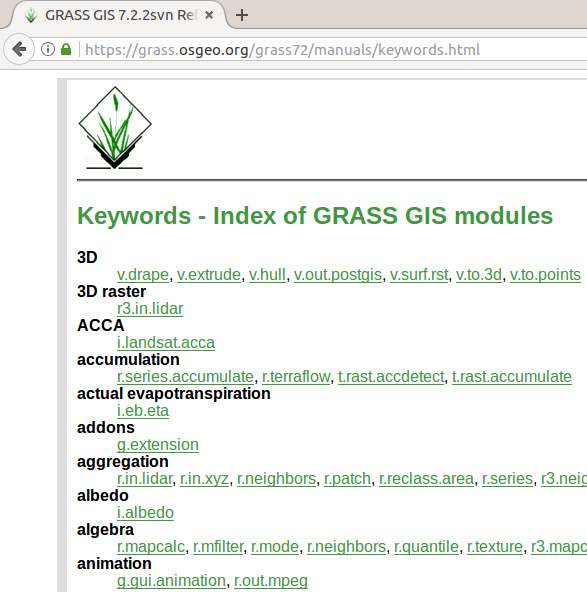
Modules and their descriptions with examples can be found the documentation. The documentation is included in the local installation and is also available online.
-
List of keywords (tags) in the online documentation
-
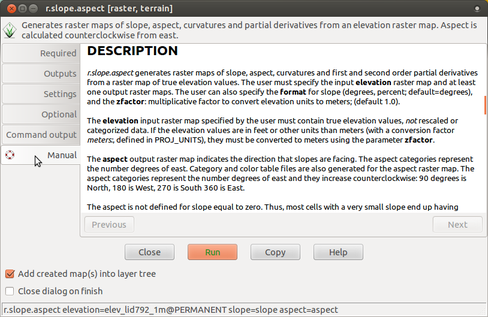
Manual page for a module is available also from the module dialog
Basic introduction to Python interface
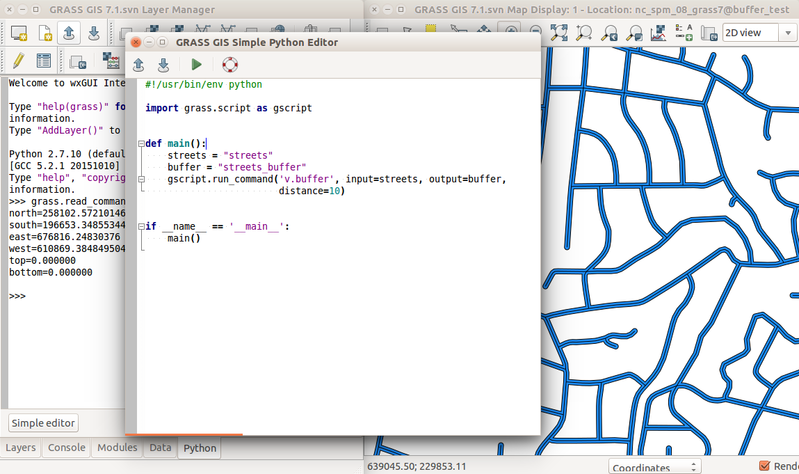
The simplest way to execute the Python code which uses GRASS GIS packages is to use Simple Python Editor integrated in GRASS GIS (accessible from the toolbar or the Python tab in the Layer Manager). Another option is to use your favorite text editor and then run the script in GRASS GIS using the main menu File → Launch script.
-
Simple Python Editor integrated in GRASS GIS
-
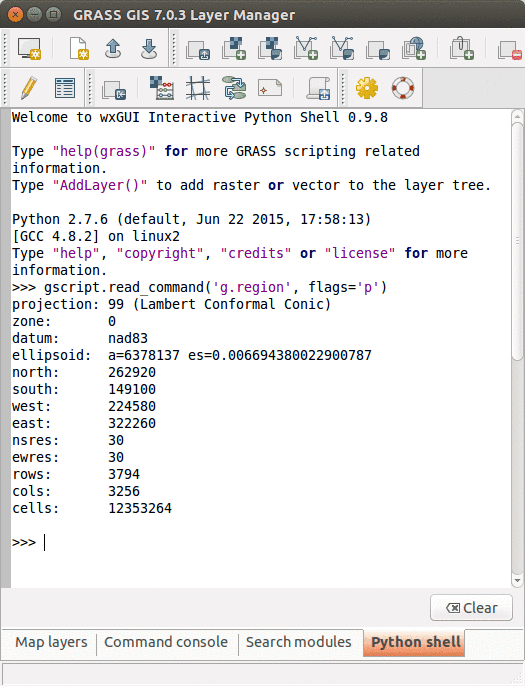
Python tab with an interactive Python shell
We will use the Simple Python Editor to run the commands. You can open it from the Python tab. When you open Simple Python Editor, you find a short code snippet. It starts with importing GRASS GIS Python Scripting Library:
import grass.script as gscript
In the main function we call g.region to see the current computational region settings:
gscript.run_command('g.region', flags='p')
Note that the syntax is similar to command line syntax (g.region -p), only the flag is specified in a parameter. Now we can run the script by pressing the Run button in the toolbar. In Layer Manager we get the output of g.region.
In this example, we set the computational extent and resolution to the raster layer ortho which would be done using g.region raster=ortho in the command line.
To use the run_command to set the computational region, replace the previous g.region command with the following line:
gscript.run_command('g.region', raster='ortho')
The GRASS GIS Python Scripting Library provides functions to call GRASS modules within Python scripts as subprocesses. All functions are in a package called grass and the most common functions are in grass.script package which is often imported import grass.script as gscript. The most often used functions include:
- script.core.run_command(): used with modules which output raster or vector data and when text output is not expected
- script.core.read_command(): used when we are interested in text output which is returned as Python string
- script.core.parse_command(): used with modules producing text output as key=value pair which is automatically parsed into a Python dictionary
- script.core.write_command(): for modules expecting text input from either standard input or file
Here we use parse_command to obtain the statistics as a Python dictionary
region = gscript.parse_command('g.region', flags='g')
print region['ewres'], region['nsres']
The results printed are the raster resolutions in E-W and N-S directions.
In the above examples, we were calling the g.region module. Typically, the scripts (and GRASS GIS modules) don't change the computational region and often they don't need to even read it. The computational region can be defined before running the script so that the script can be used with different computational region settings.
The library also provides several convenient wrapper functions for often called modules, for example script.raster.raster_info() (wrapper for r.info), script.core.list_grouped() (one of the wrappers for g.list), and script.core.region() (wrapper for g.region).
When we want to run the script again, we need to either remove the created data beforehand using g.remove or we need to tell GRASS GIS to overwrite the existing data. This can be done adding overwrite=True as an additional argument to the function call for each module or we can do it globally using os.environ['GRASS_OVERWRITE'] = '1' (requires import os).
Finally, you may have noticed the first line of the script which says #!/usr/bin/env python. This is what Linux, Mac OS, and similar systems use to determine which interpreter to use. If you get something line [Errno 8] Exec format error, this line is probably incorrect or missing.
Decide if to use GUI, command line, Python, or Jupyter Notebook
- GUI: can be combined anytime with command line (recommended, especially for beginners, combine with copy and paste to Console)
- command line: can be any time combined with the GUI, most of the following instructions will be for command line (but can be easily transfered to GUI or Python), both system command line and Console tab in GUI will work well
- Python: you will need to change the syntax to Python; use snippets from this static notebook
- Jupyter Notebook locally: this is recommended only if you are using OSGeoLive or if you are on Linux and are familiar with Jupyter and GRASS GIS; download notebook from github.com/wenzeslaus/Notebook-for-processing-point-clouds-in-GRASS-GIS
- Jupyter Notebook online: link distributed by the instructor (backup option if other options fail)
Binning of the point cloud
-
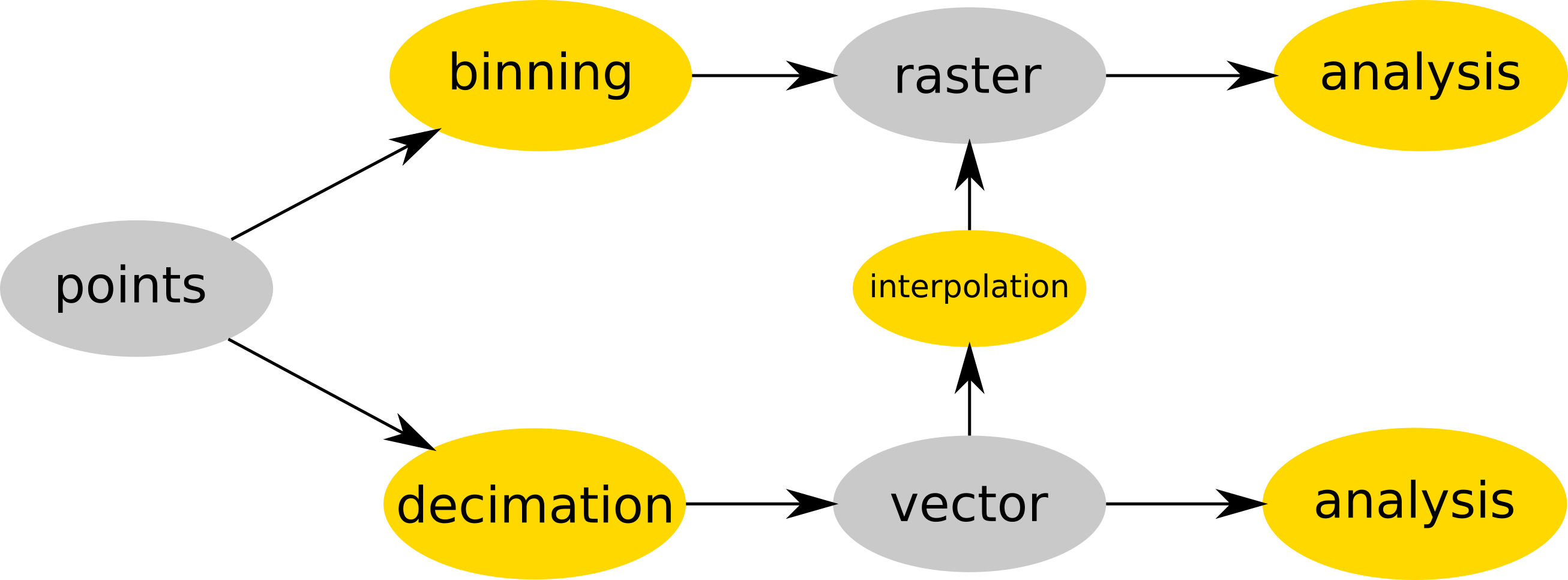
Point cloud is either binned into a raster (e.g. r.in.lidar) and then analyzed as raster or optionally decimated (e.g. using v.in.lidar), converted to vector (still using v.in.lidar) and then interpolated (e.g. v.surf.rst) into raster or analyzed as a vector (e.g. v.vect.stats). Data are in gray, processes in yellow.
-
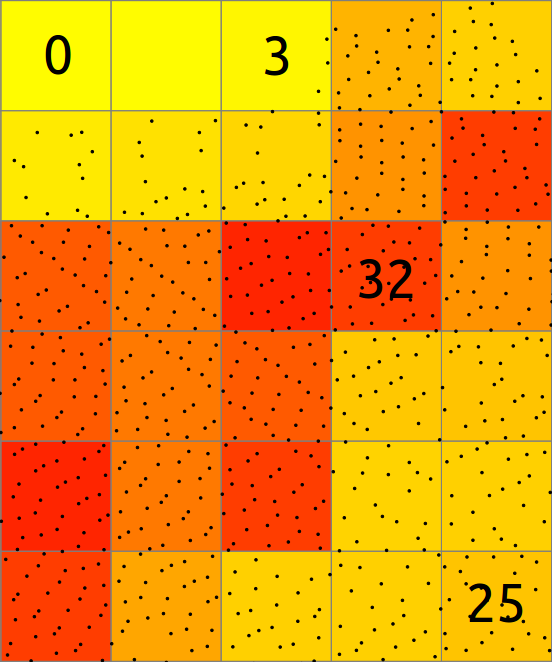
In basic case, binning of points into a 2D raster consists of counting the number of points falling into each cell. The resulting cell value is then count of points in that cell.
-
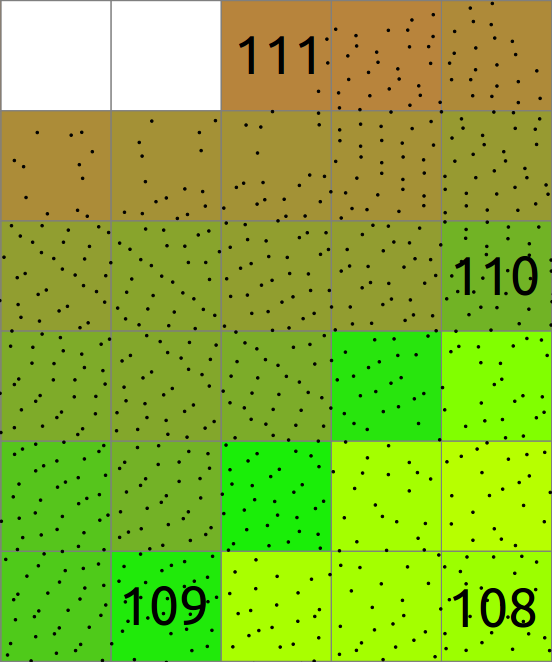
In general binning involves also values associated with the points and computes statistics on these values. Here the mean of Z coordinates of all the point in each cell is computed and stored in the raster. The cells without any points are NULL (NoData) shown in white here.
Fastest way to analyze basic properties of a point cloud is to use binning and create a raster map. We will now use r.in.lidar to create point count (point density) raster. At this point, we don't know the spatial extent of the point cloud, so we cannot set computation region ahead, but we can tell the r.in.lidar module to determine the extent first and use it for the output using the -e flag. We are using a coarse resolution to maximize the speed of the process. Additionally, the -r flag sets the computational region to match the output.
r.in.lidar input=nc_tile_0793_016_spm.las output=count_10 method=n -e -n resolution=10
Now we can see the distribution pattern, but let's examine also the numbers (in GUI using right click on the layer in Layer Manager and then Metadata or using r.info directly):
r.info map=count_10
We can do a quick check of some of the values using query tool in the Map Display. Since there is a lot of points per one cell, we can use a finer resolution:
r.in.lidar input=nc_tile_0793_016_spm.las output=count_1 method=n -e -n resolution=1
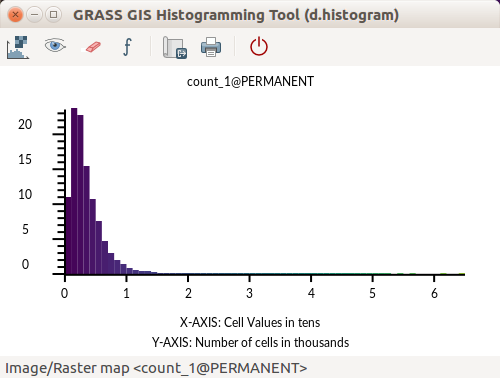
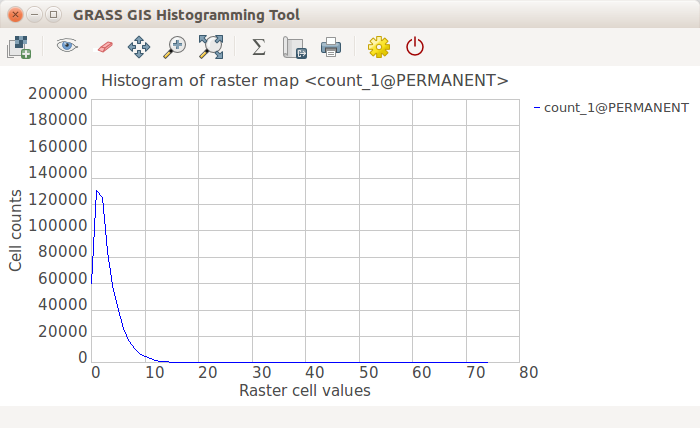
Look at the distribution of the values using a histogram. Histogram is accessible from the context menu of a layer in Layer Manager, from Map Display toolbar Analyze map button or using the d.histogram module (d.histogram map=count_1).
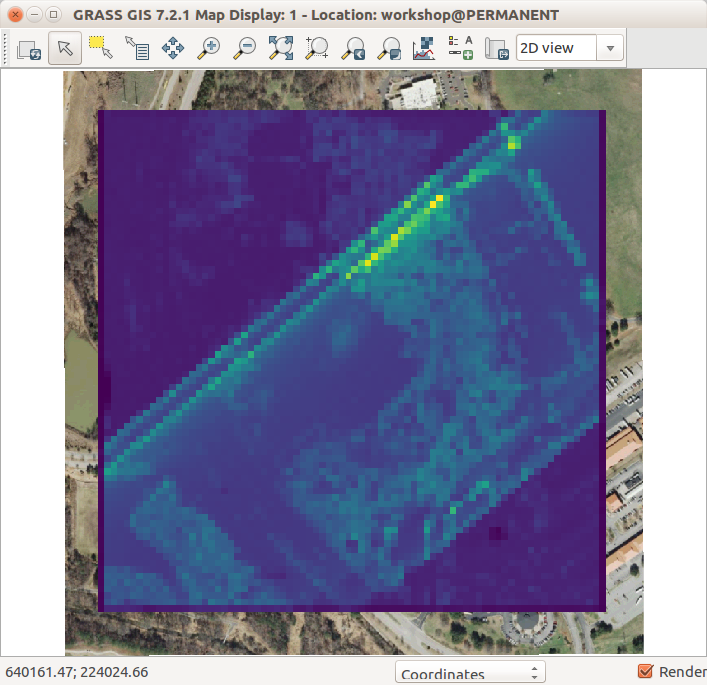
-
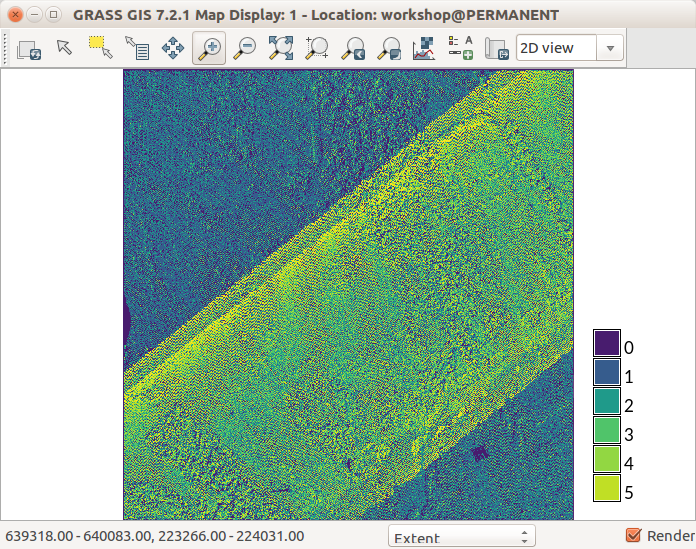
Lidar point count per cell using r.in.lidar with 10 m cells (
resolution=10) and-eflag -
GRASS GIS Histogramming Tool (based on d.histogram)
-
GRASS GIS Histogramming Tool (based on wxPython)
Now we have appropriate number of points in most of the cells and there are no density artifacts around the edges, so we can use this raster as the base for extent and resolution we will use from now on.
g.region raster=count_1 -p
However, this region has a lot of cells and some of them would be empty, esp. when we start filtering the point cloud. To account for that, we can modify just the resolution of the computational region:
g.region res=3 -ap
Use binning to obtain the digital surface model (DSM) (resolution and extent are taken from the computational region):
r.in.lidar input=nc_tile_0793_016_spm.las output=binned_dsm method=max
To understand what the map shows, compute statistics using r.report (Report raster statistics in the layer context menu):
r.report map=binned_dsm units=c
This shows that there is an outlier (at around 600 m). Change the color table to histogram equalized (-e flag) to see the contrast in the rest of the map (using the viridis color table):
r.colors map=binned_dsm color=elevation -e
Let's check the outliers also using minimum (which is method=min, we already used method=max for DSM):
r.in.lidar input=nc_tile_0793_016_spm.las output=minimum method=min
Compare this with range and mean of the values in the ground raster and decide what is the permissible range.
r.report map=minimum units=c
Again, to see the data, we can use histogram equalized color table:
r.colors map=minimum color=elevation -e
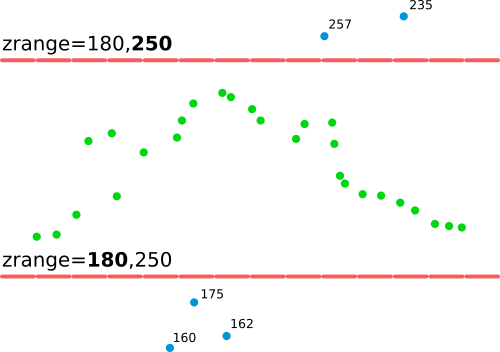
zrange parameter filters out points which don't fall into the specified range of Z valuesNow when we know the outliers and the expected range of values (60-200 m seems to be a safe and but broad enough range), use the zrange parameter to filter out any possible outliers (don't be confused with the intensity_range parameter) when computing a new DSM:
r.in.lidar input=nc_tile_0793_016_spm.las output=binned_dsm_limited method=max zrange=60,200
Interpolation
Now we will interpolate a digital surface model (DSM) and for that we can increase resolution to obtain as much detail as possible (we could use 0.5 m, i.e. res=0.5, for high detail or 2 m, i.e. res=2 -a, for speed):
g.region raster=count_1 -p
Before interpolating, let's confirm that the spatial distribution of the points allows us to safely interpolate. We need to use the same filters as we will use for the DSM itself:
r.in.lidar input=nc_tile_0793_016_spm.las output=count_dsm method=n return_filter=first zrange=60,200
First check the numbers:
r.report map=count_dsm units=h,c,p
Then check the spatial distribution. For that we will need to use histogram equalized color table (legend can be limited just to a specific range of values: d.legend raster=count_interpolation range=0,5).
r.colors map=count_dsm color=viridis -e
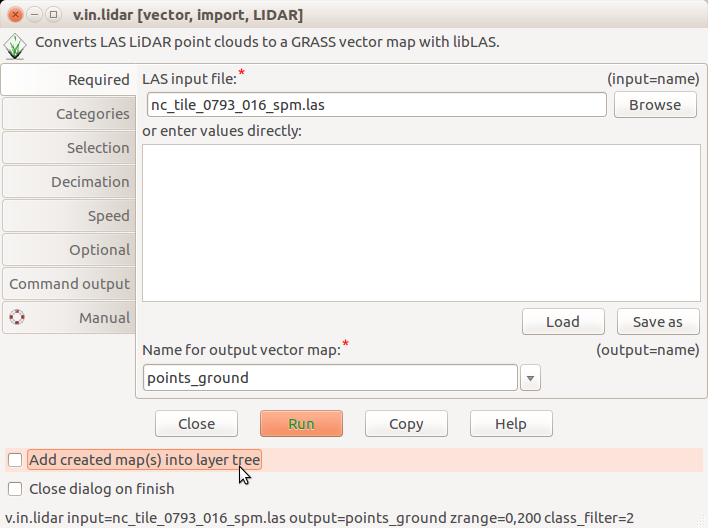
First we import LAS file as vector points, we keep only first return points and limit the import vertically to avoid using outliers found in the previous step. Before running it, uncheck Add created map(s) into layer tree in the v.in.lidar dialog if you are using GUI.
v.in.lidar -bt input=nc_tile_0793_016_spm.las output=first_returns return_filter=first zrange=60,200
-
Unchecked Add created map(s) into layer tree in v.in.lidar
-
Point count at fine resolution with histogram equalized viridis color table and legend with a limited range
Then we interpolate:
v.surf.rst input=first_returns elevation=dsm tension=25 smooth=1 npmin=80
Now we could visualize the resulting DSM by computing shaded relief with r.relief or by using 3D view (see the following sections).
-
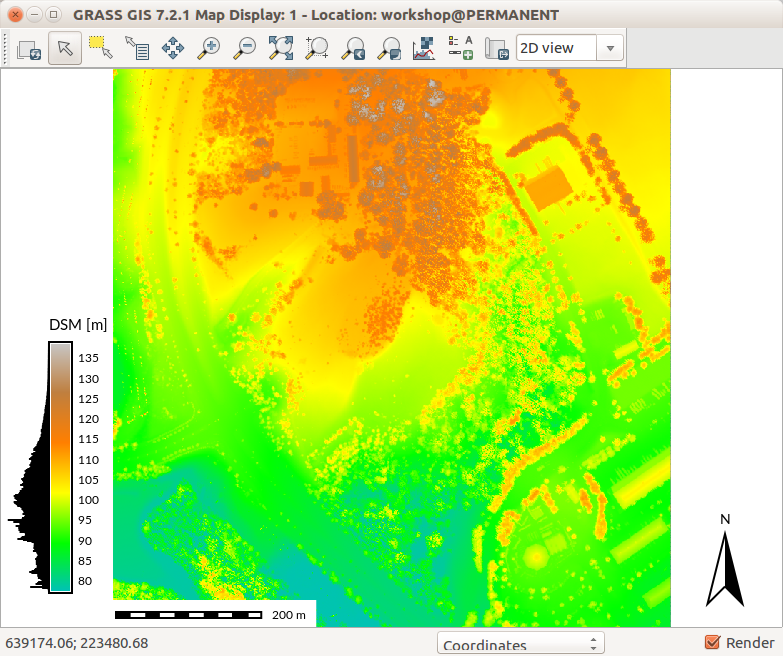
DSM with legend and histogram.
Terrain analysis
Set the computational region based on an existing raster map:
g.region raster=count_1 -p
Check if the point density is sufficient (ground is class number 2, resolution and extent are taken from the computational region):
r.in.lidar input=nc_tile_0793_016_spm.las output=count_ground method=n class_filter=2 zrange=60,200
Import points (the -t flag disables creation of attribute table and the -b flag disables building of topology; uncheck Add created map(s) into layer tree):
v.in.lidar -bt input=nc_tile_0793_016_spm.las output=points_ground class_filter=2 zrange=60,200
-
Map Display with legend created using d.legend, r.in.lidar dialog, ground point density pattern in the background
-
Unchecked Add created map(s) into layer tree in v.in.lidar
The interpolation will take some time, but we can set the computational region to a smaller area to save some time before we determine what are the best parameters for interpolation. This can be done in the GUI from the Map Display toolbar or here just quickly using the prepared coordinates:
g.region n=223751 s=223418 w=639542 e=639899 -p
-
Simple ways to set computational region extent from GUI. On the left, set region to match raster map. On the right, select the highlighted option and then set region by drawing rectangle. Then zoom to computational region.
-
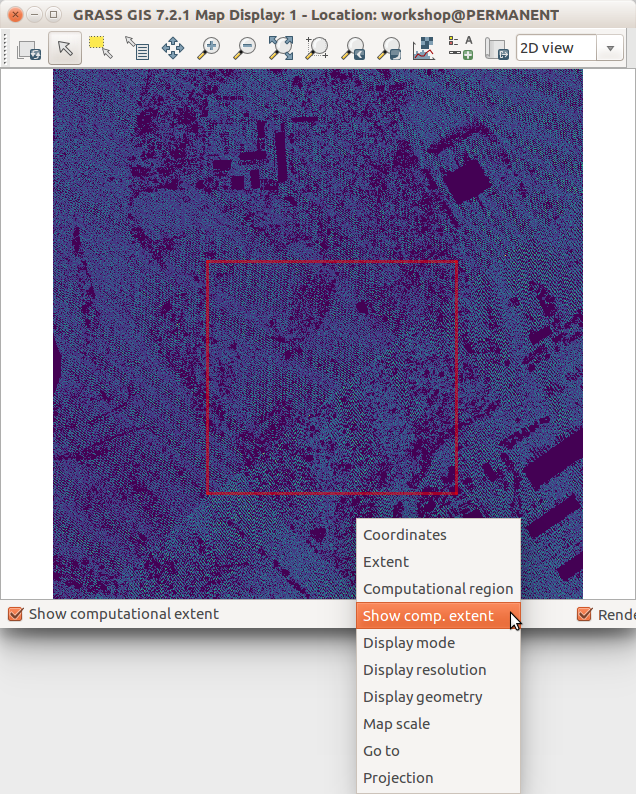
Show the current computational region extent in the Map Display
Now we interpolate the ground surface using regularized spline with tension (implemented in v.surf.rst) and at the same time we also derive slope, aspect and curvatures (the following code is one long line):
v.surf.rst input=points_ground tension=25 smooth=1 npmin=100 elevation=terrain slope=slope aspect=aspect pcurvature=profile_curvature tcurvature=tangential_curvature mcurvatur=mean_curvature
When we examine the results, especially the curvatures curvatures show a pattern which may be caused by some problems with the point cloud collection. We decrease the tension which will cause the surface to hold less to the points and we increase the smoothing. Since the raster map called range already exists, use the overwrite flag, i.e. --overwrite in the command line or checkbox in GUI to replace the existing raster by the new one. We use the --overwrite flag shortened to just --o.
v.surf.rst input=points_ground tension=20 smooth=5 npmin=100 elevation=terrain slope=slope aspect=aspect pcurvature=profile_curvature tcurvature=tangential_curvature mcurvatur=mean_curvature --o
When we are satisfied with the result, we get back to our desired raster extent:
g.region raster=count_1 -p
And finally interpolate the ground surface in the full extent:
v.surf.rst input=points_ground tension=20 smooth=5 npmin=100 elevation=terrain slope=slope aspect=aspect pcurvature=profile_curvature tcurvature=tangential_curvature mcurvatur=mean_curvature --o
Compute shaded relief:
r.relief input=terrain output=relief
Now combine the shaded relief raster and the elevation raster. This can be done in several ways. In GUI by changing opacity of one of them. A better result can be obtained using the r.shade module which combines the two rasters and creates a new one. Finally, this can be done on the fly without creating any raster using the d.shade module. The module can be used from GUI through toolbar or using the Console tab:
d.shade shade=relief color=terrain
Now, instead of using r.relief, we will use r.skyview:
r.skyview input=terrain output=skyview ndir=8 colorized_output=terrain_skyview
Combine the terrain and skyview also on the fly:
d.shade shade=skyview color=terrain
Analytical visualization based on shaded relief:
r.shaded.pca input=terrain output=pca_shade
Local relief model:
r.local.relief input=terrain output=lrm shaded_output=shaded_lrm
Pit density:
r.terrain.texture elevation=terrain thres=0 pitdensity=pit_density
Finally, we use automatic detection of landforms (using 50 m search window):
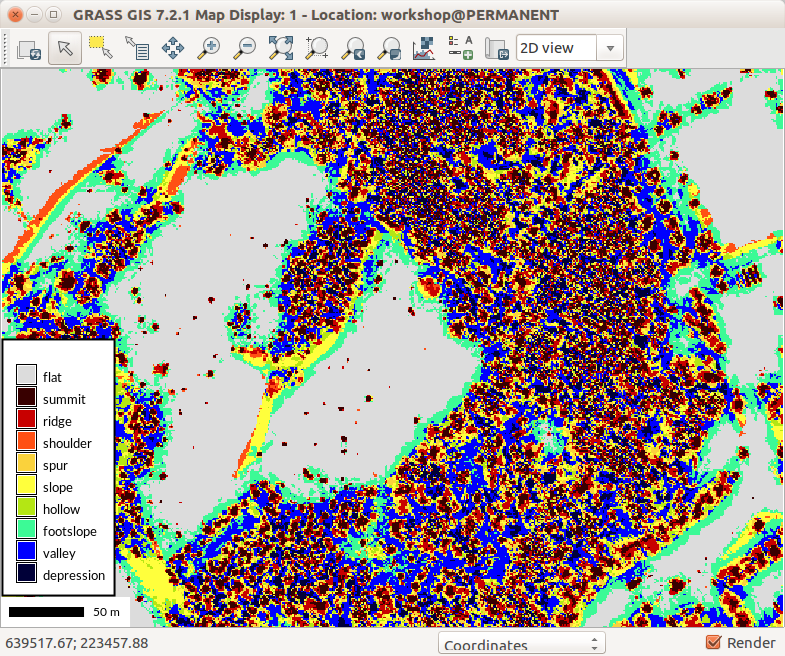
r.geomorphon -m elevation=terrain forms=landforms search=50
-
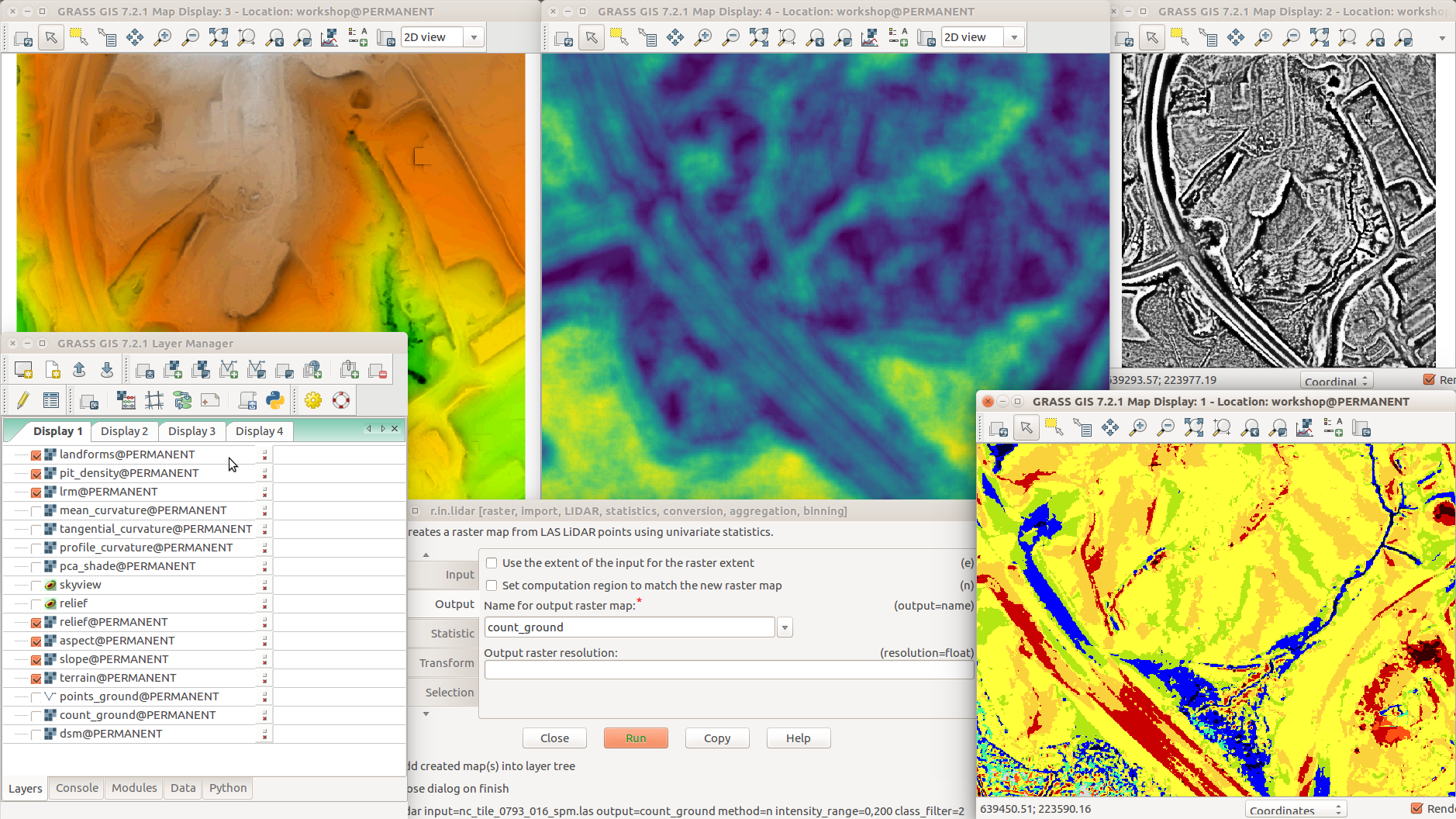
Different terrain analyses and visualizations in multiple Map Displays
Vegetation analysis

zrange parameter filters out points which don't fall into the specified range of Z valuesAlthough for many vegetation-related application, coarser resolution is more appropriate because more points are needed for the statistics, we will use just:
g.region raster=count_1
We use all the points (not using the classification of the points), but with the Z filter to get the range of heights in each cell:
r.in.lidar input=nc_tile_0793_016_spm.las output=range method=range zrange=60,200
We use all the points, but with the Z filter to get the range of heights in each cell:
r.mapcalc "vegetation_by_range = if(range > 2, 1, null())"
Or lower res to avoid gaps (and possible smoothing during their filling).
-
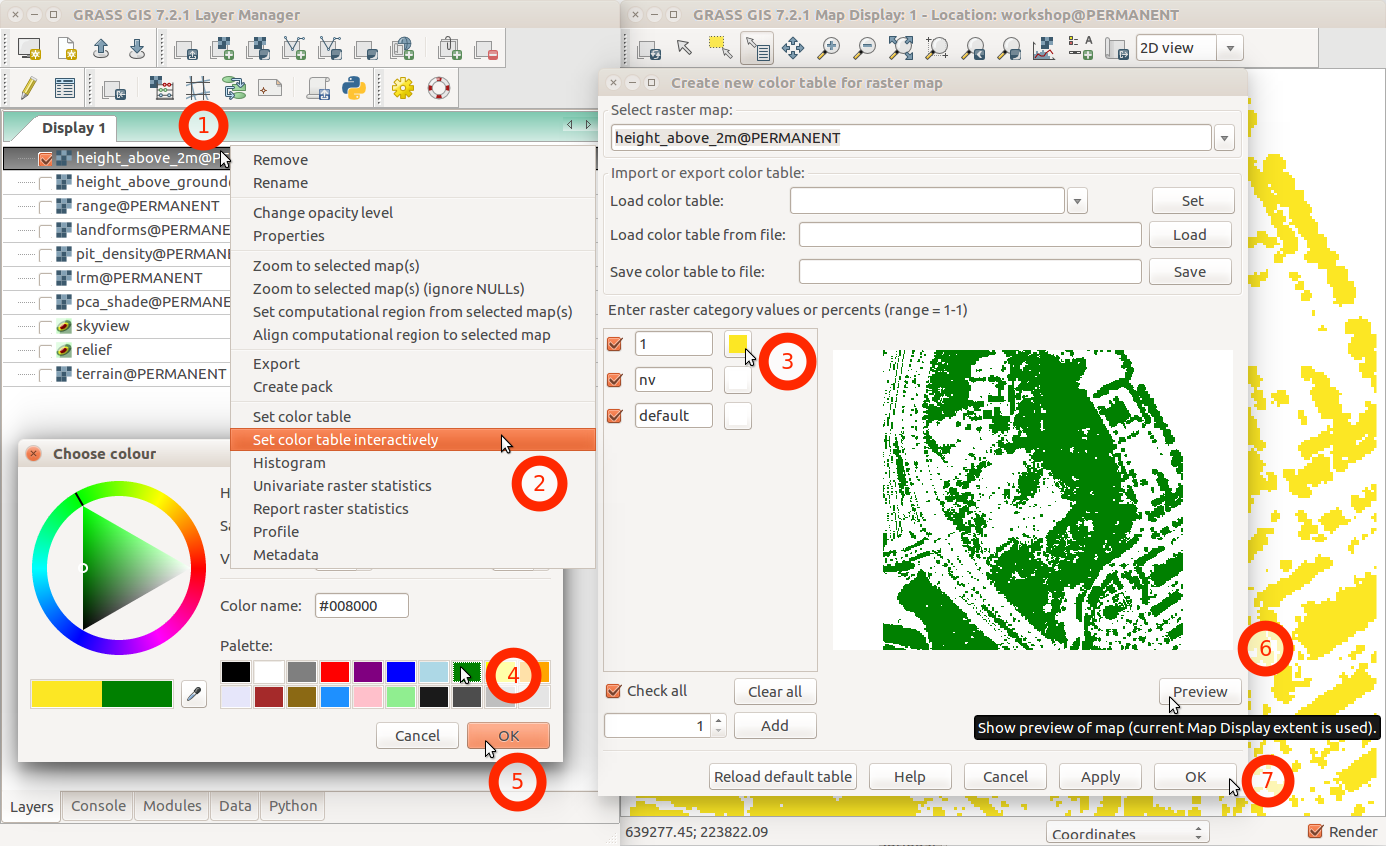
Change color table for a raster map from layer context menu using Set color table interactively.
r.in.lidar -d input=nc_tile_0793_016_spm.las output=height_above_ground method=max base_raster=terrain zrange=0,100
Now get all the areas which are higher than 2 m.
In case we want to preserve the elevation in the output,
we could use if(height_above_ground > 2, height_above_ground, null()) in the expression,
but we want just the areas, so we use constant (1).
r.mapcalc "above_2m = if(height_above_ground > 2, 1, null())"
Use r.grow to extend the patches and fill in the holes:
r.grow input=above_2m output=vegetation_grow
We consider the result to represent the vegetated areas. We clump (connect) the individual cells into patches using r.clump):
r.clump input=vegetation_grow output=vegetation_clump
Some of the patches are very small. Using r.area we remove all patches smaller than the given threshold:
r.area input=vegetation_clump output=vegetation_by_height lesser=100
Now convert these areas to vector:
r.to.vect -s input=vegetation_by_height output=vegetation_by_height type=area
So far we were using elevation of the points, now we will use intensity.
The intensity is used by r.in.lidar when -j (or -i) flag is provided:
r.in.lidar input=nc_tile_0793_016_spm.las output=intensity zrange=60,200 -j
With this high resolution, there are some cells without any points, so we use r.fill.gaps to fill these cells (r.fill.gaps also smooths the raster as part of the gap-filling process).
r.fill.gaps input=intensity output=intensity_filled uncertainty=uncertainty distance=3 mode=wmean power=2.0 cells=8
Grey scale color table is more appropriate for intensity:
r.colors map=intensity_filled color=grey
There are some areas with very high intensity, so to visually explore the other areas, we use histogram equalized color table:
r.colors -e map=intensity_filled color=grey
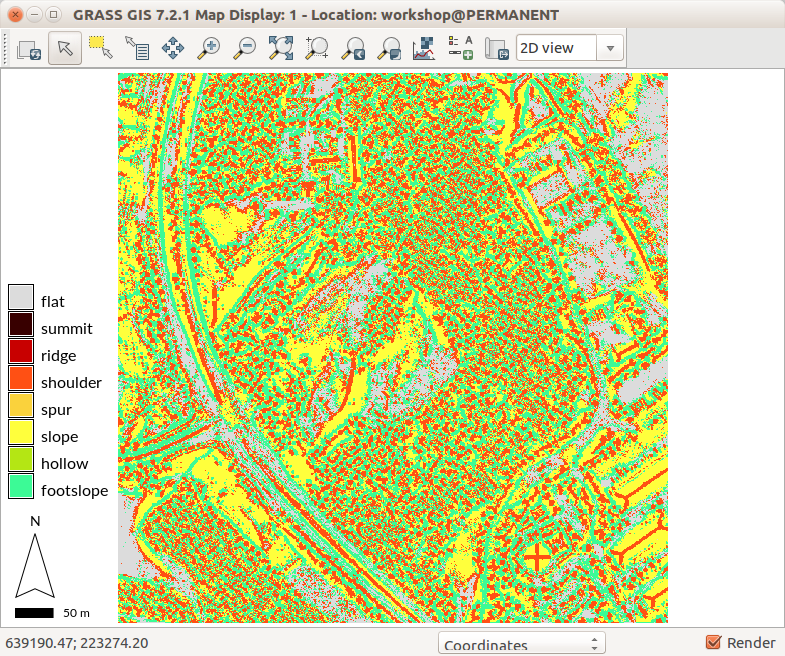
Let's use r.geomorphon again, but now with DSM and different settings. The following settings shows building roof shapes:
r.geomorphon elevation=dsm forms=dsm_forms search=7 skip=4
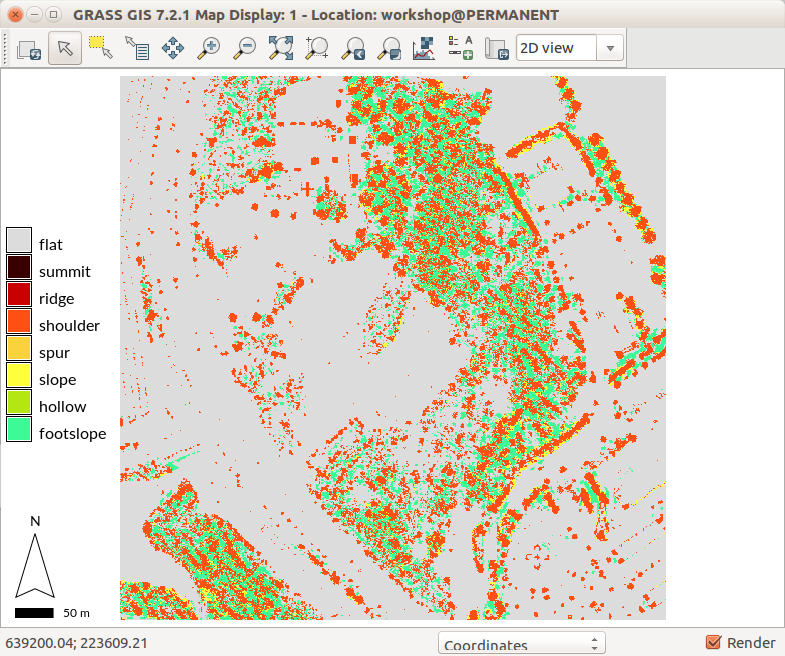
Different settings, especially higher flatness threshold (flat parameter) shows forested areas as combination of footslopes, slopes, and shoulders. Individual trees are represented as shoulders.
r.geomorphon elevation=dsm forms=dsm_forms search=12 skip=8 flat=10 --o
Lowering the skip parameter decreases generalization and brings in summits which often represents tree tops:
r.geomorphon elevation=dsm forms=dsm_forms search=12 skip=2 flat=10 --o
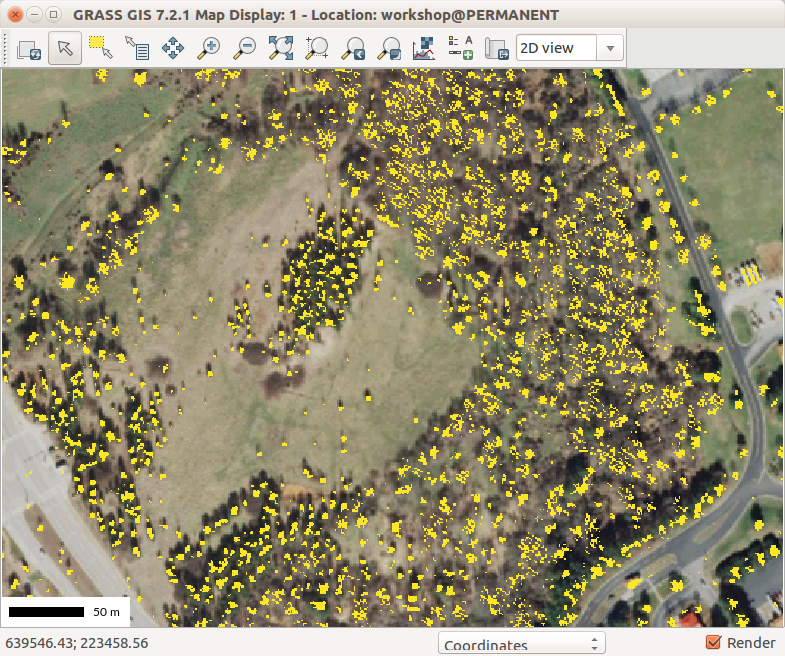
Now we extract summits using r.mapcalc raster algebra expression if(dsm_forms==2, 1, null()).
r.mapcalc "trees = if(dsm_forms==2, 1, null())"
-
r.geomorphon settings which shows well shapes of roofs
-
Different r.geomorphon settings showing vegetated areas
-
More common setting of r.geomorphon resulting in detailed description of DSM shapes
-
Selected peaks from r.geomorphon showing positions of trees (tree crowns)
Bonus tasks
3D visualization of rasters
We can explore our study area in 3D view (use image on the right if clarification is needed for below steps):
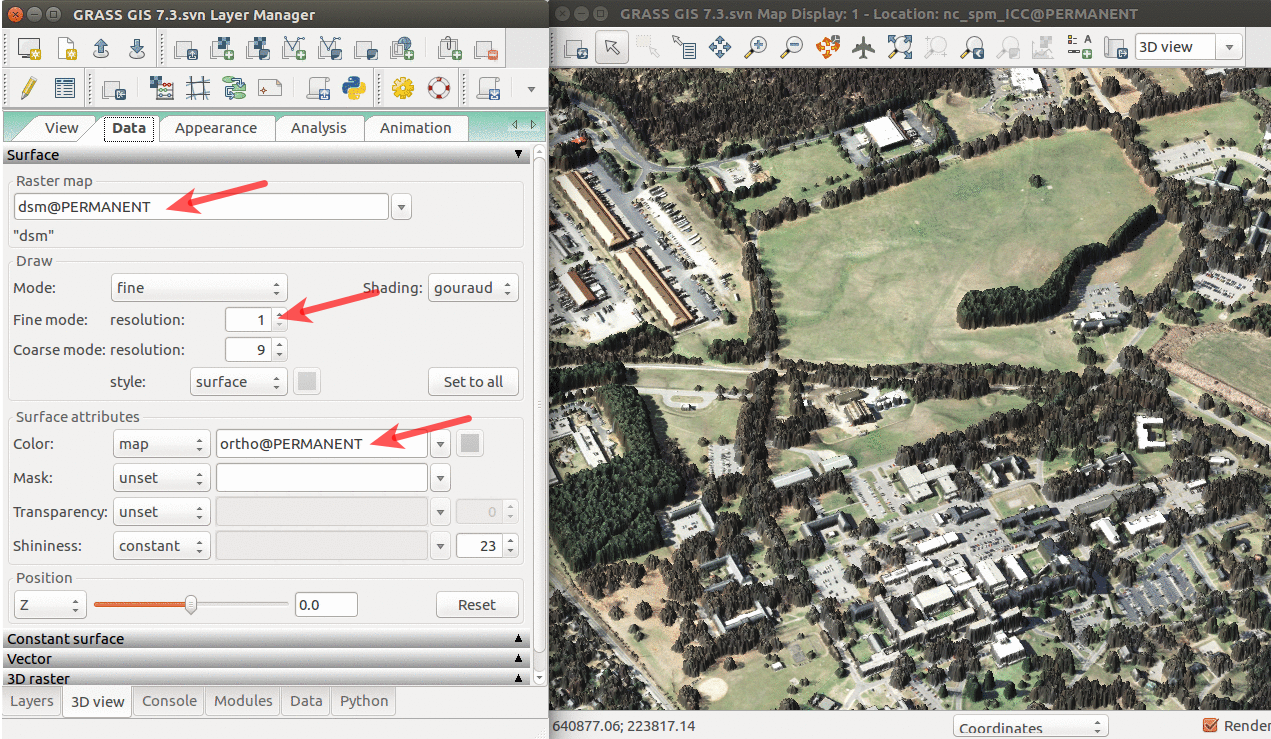
- Add raster dsm and uncheck or remove any other layers. Note that unchecking (or removing) any other layers is important because any layer loaded in Layers is interpreted as a surface in 3D view.
- Switch to 3D view (in the right corner of Map Display).
- Adjust the view (perspective, height).
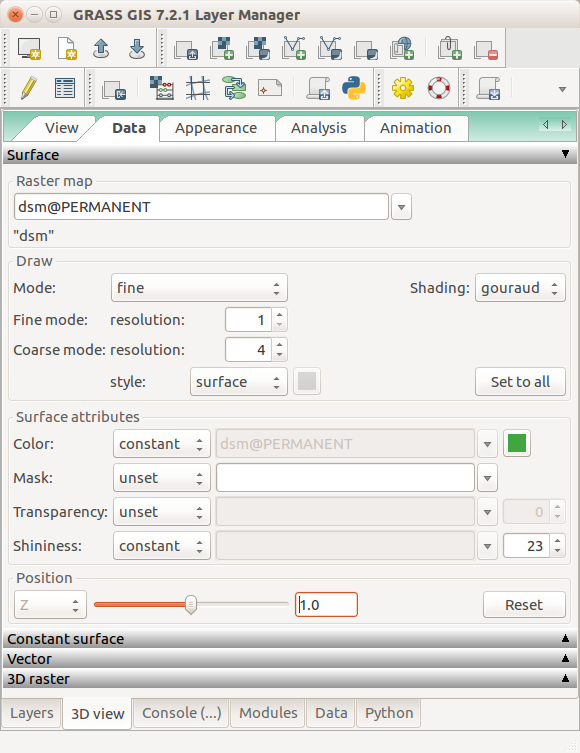
- In Data tab, set Fine mode resolution to 1 and set ortho as the color of the surface (the orthophoto is draped over the DSM).
- Go back to View tab and explore the different view directions using the green puck.
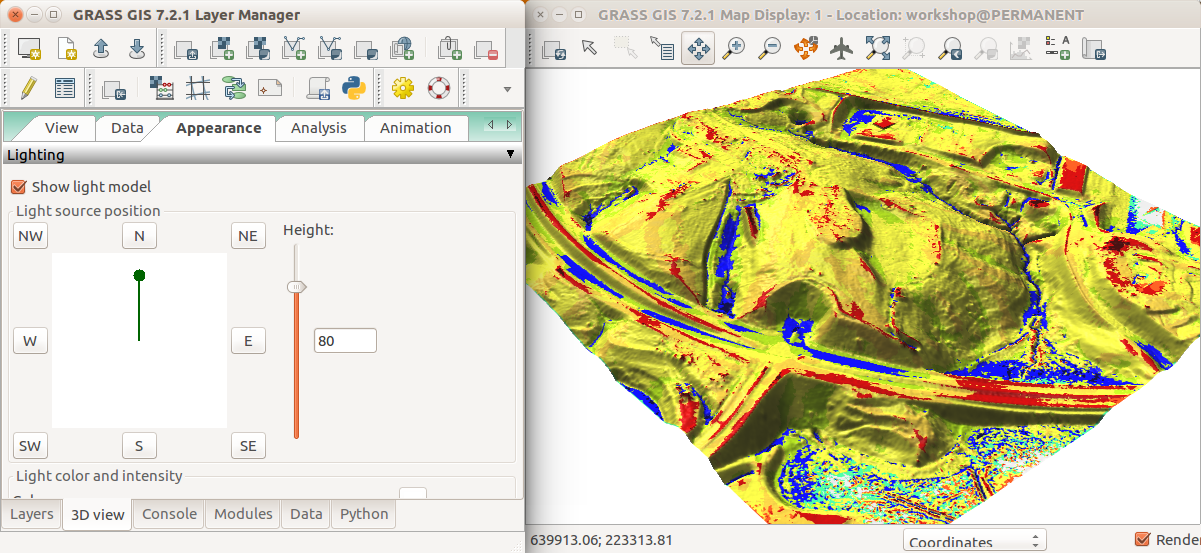
- Go to Appearance tab and change the light conditions (lower the light height, change directions).
- Try also using the landforms raster as the color of the surface.
- When finished, switch back to 2D view.
-
3D visualization of DSM with orthophoto draped over: arrows pointing to the name of the surface, resolution used for its display, and raster used as its color.
-
Landforms over the DSM in 3D: The light source position is changed in the Appearance tab.
Analytical visualization of rasters in 3D
We can explore our study area in 3D view (use image on the right if clarification is needed for below steps):
- Add rasters terrain and dsm and uncheck or remove any other layers. (Any layer in Layers will be interpreted as surface in 3D view.)
- Switch to 3D view (in the right corner of Map Display).
- Adjust the view (perspective, height). Set z-exaggeration to 1.
- In Data tab, in Surface, set Fine mode resolution to 1 for both rasters.
- Set a different color for each surface.
- For the dsm, set the position in Z direction to 1. This is a position relative the actual position of the raster. This small offset will help is see relation in between the terrain and the DSM surfaces.
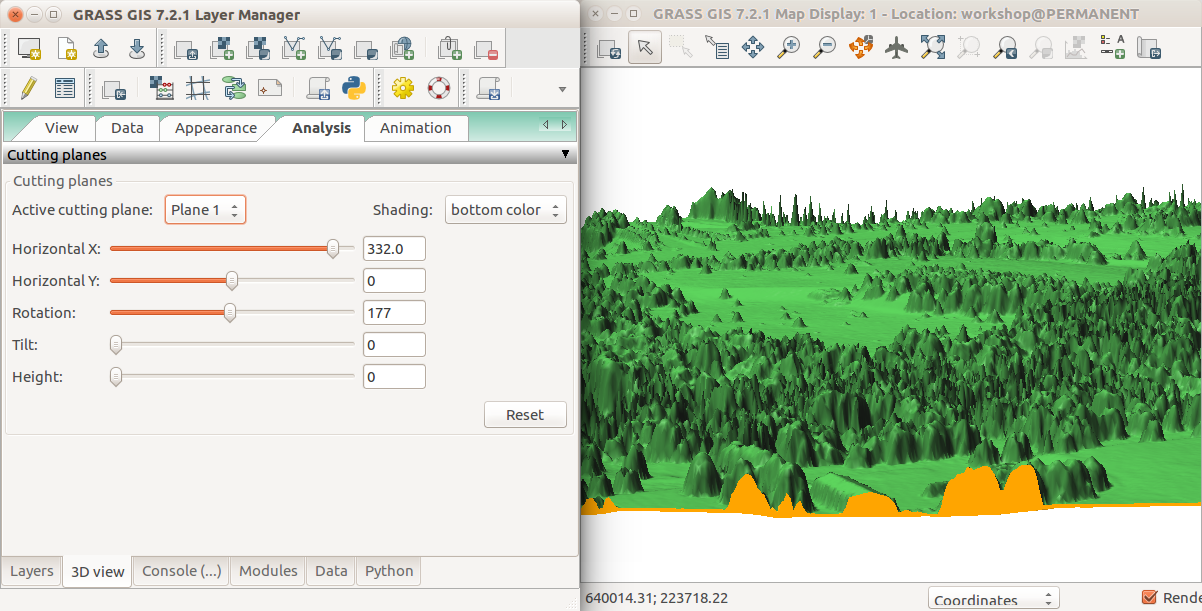
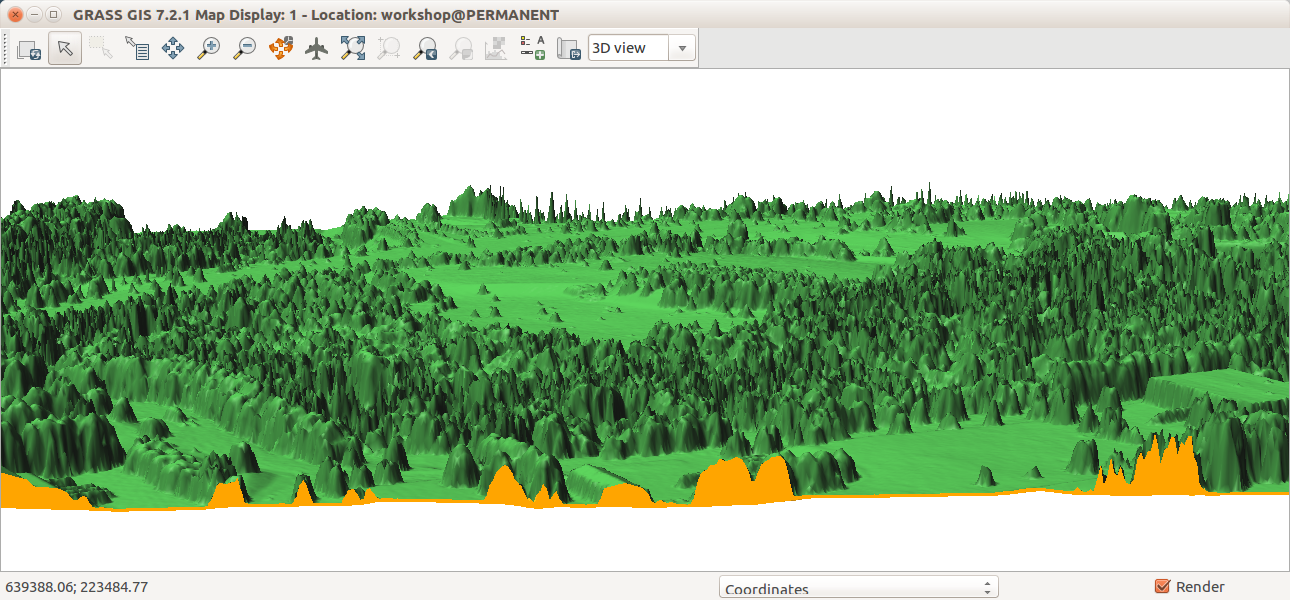
- Go to Analysis tab and activate the first cutting plane. Set Shading to bottom color to get the bottom color to color the space between the terrain and DSM.
- Start sliding the sliders for X, Y and Rotation.
- Go back to View tab in case you want to change the view.
- When finished, switch back to 2D view.
-
In the Data tab of 3D view, set the same fine mode resolution for both surfaces. Choose different colors. Set (relative) position of the DSM to 1 m (above its actual position).
-
In the Analysis tab of 3D view, activate cutting plane, set shading to bottom color and start sliding the sliders for X, Y and rotation.
-
To see longer part of the transect, stretch the window horizontally.
Classify ground and non-ground points
UAV point clouds usually require classification of ground points (bare earth) when we want to create ground surface.
Import all the points using the v.in.lidar command in the previous section, unless you already have them. Since the metadata about projection are wrong, we use the -o flag to skip the projection consistency check.
r.in.lidar -e -n -o input=nc_uav_points_spm.las output=uav_density_05 method=n resolution=0.5
Set a small computational region to make all the computations faster (skip this to operate in the whole area):
g.region n=219415 s=219370 w=636981 e=637039
Import the points using the v.in.lidar module but limit the extent just to computational region (-r flag), do not build topology (-b flag), do not create attribute table (-t flag), and do not assign categories (ids) to points (-c flag). There is more points than we need for interpolating the the resolution 0.5 m, so we will decimate (thin) the point cloud during import removing 75% of the points using preserve=4 (which uses count-based decimation which assumes spatially uniform distribution of the points).
v.in.lidar -t -c -b -r -o input=nc_uav_points_spm.las output=uav_points preserve=4
Then use v.lidar.mcc to classify ground and non-ground points:
v.lidar.mcc input=points_all ground=mcc_ground nonground=mcc_nonground
Interpolate the ground surface:
v.surf.rst input=mcc_ground tension=20 smooth=5 npmin=100 elevation=mcc_ground
If you are using the UAV data, you can extract the RGB values from the points. First, import the points again, but this time with all the attributes:
v.in.lidar -r -o input=nc_uav_points_spm.las output=uav_points preserve=4
Then extract the RGB values (stored in attribute columns) into separate channels (rasters):
v.to.rast input=uav_points output=red use=attr attr_col=red v.to.rast input=uav_points output=green use=attr attr_col=green v.to.rast input=uav_points output=blue use=attr attr_col=blue
And then set the color table to grey:
r.colors red color=grey r.colors green color=grey r.colors blue color=grey
These channels could be used for some remote sensing tasks. Now we will just visualize them using d.rgb:
d.rgb red=red green=green blue=blue
If you are using the lidar data for this part, you can compare the new DEM with the previous dataset:
r.mapcalc "mcc_lidar_differece = ground - mcc_ground"
Set the color table:
r.colors map=mcc_lidar_differece color=difference
3D visualization of point clouds
Unless you have already done that, familiarize yourself with the 3D view by doing the exercise above. Then import all points but this time do not add them to the Layer Manager (there is a check box at the bottom of the module dialog).
v.in.lidar -obt input=nc_tile_0793_016_spm.las output=points_all zrange=60,200
Now we can explore point clouds in 3D view:
- Uncheck or remove any other layers. (Any layer in Layer Manager is interpreted as surface in 3D view.)
- Switch to 3D view (in the right corner of Map Display).
- When 3D is active, Layer Manager has a new icon for default settings of the 3D which we need to open and change.
- In the settings, change settings for default point shape from sphere to X.
- Then add the points vector.
- You may need to adjust the size of the points if they are all merged together.
- Adjust the view (perspective, height).
- Go back to View tab and explore the different view directions using the green puck.
- Go to Appearance tab and change the light conditions (lower the light height, change directions).
- When finished, switch back to 2D view.
-
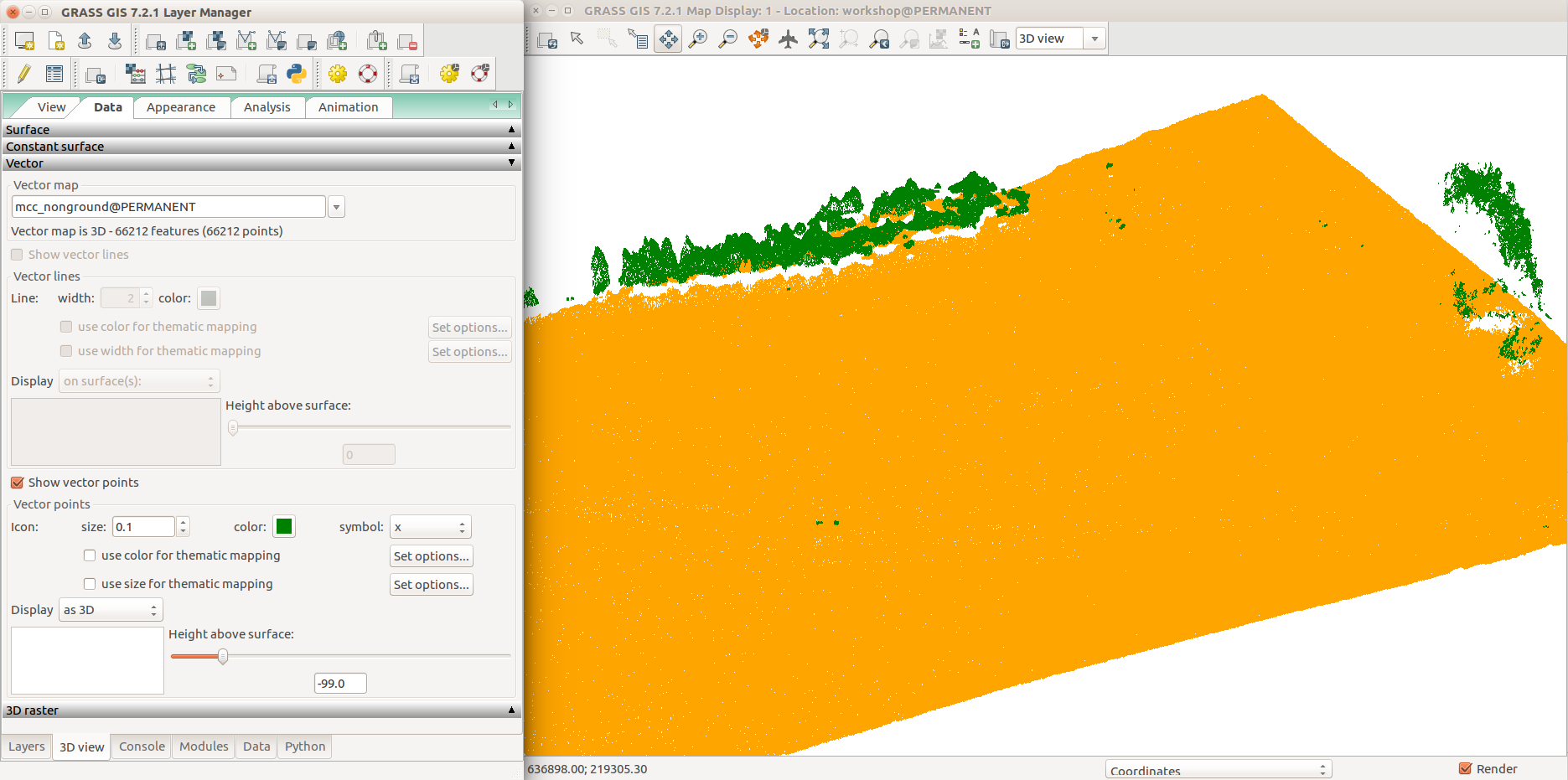
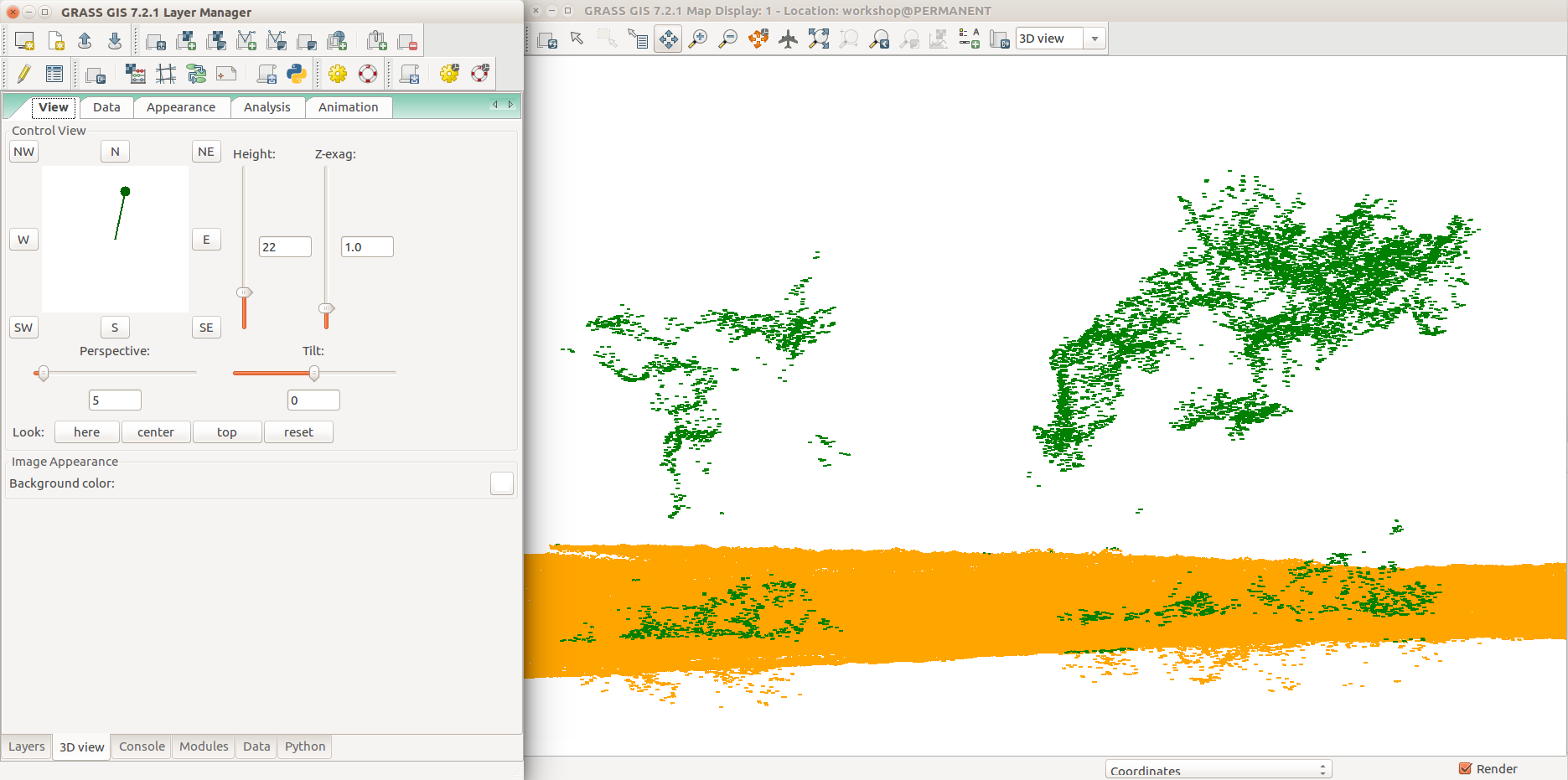
UAV point cloud classified to group points (orange) and non-ground points cloud (green).
-
Detail showing trees and outliers.
Alternative DSM creation
First returns not always represent top of the canopy, but they can simply represent first hit somewhere in the canopy. To account for this we create a raster with maximum for each cell instead of importing the raw points:
r.in.lidar input=nc_tile_0793_016_spm.las output=maximum method=maximum return_filter=first
The raster has many missing cells, that's why we want to interpolate, but at the same time, we also reduced the number of points by replacing all points in one cells just by this one cell. We can consider the cell to represent one point in the middle of the cell and use r.to.vect to create points from these cells:
r.to.vect input=maximum output=points_for_dsm type=point -b
Now we have a decimated (thinned) point cloud, decimated using binning into raster. We interpolate these points to get the DSM:
v.surf.rst input=first_returns elevation=dsm tension=25 smooth=1 npmin=80
Explore layers of vegetation in a 3D raster
Set the top and bottom of the computational region to match the height of vegetation (we are interested in vegetation from 0 m to 30 m above ground).
g.region -p3 b=0 t=30
Similarly to 2D, we can perform the binning also in three dimensions using 3D rasters. This is implemented in a module called r3.in.lidar:
r3.in.lidar -d input=nc_tile_0793_016_spm.las n=count sum=intensity_sum mean=intensity proportional_n=prop_count proportional_sum=prop_intensity base_raster=terrain
Convert the horizontal layers (slices) of 3D raster into 2D rasters:
r3.to.rast input=prop_count output=slices_prop_count
Open a second Map Display and add all the created rasters. Then set the a consistent color table to all of them.
r.colors slices_prop_count_001,slices_prop_count_002,... color=viridis
Now open the Animation Tool and add all to 2D rasters there and use slider to explore the layers.
Optimizations, troubleshooting and limitations
Speed optimizations:
- Rasterize early.
- For many use cases, there is less cells than points. Additionally, rasters will be faster, e.g. for visualization, because raster has a natural spatial index. Finally, many algorithms simply use rasters anyway.
- If you cannot rasterize, see if you can decimate (thin) the point cloud (using v.in.lidar, r.in.lidar + r.to.vect, v.decimate).
- Binning (e.g. r.in.lidar, r3.in.lidar) is much faster than interpolation and can carry out part of the analysis.
- Faster count-based decimation performs often the same on a given point cloud for terrain as slower grid-based decimation (Petras et al. 2016).
- r.in.lidar
- choose computation region extent and resolution ahead
- have enough memory to avoid using percent option (high memory usage versus high I/O)
- v.in.lidar
- -r limit import to computation region extent
- -t do not create attribute table
- -b do not build topology (applicable to other modules as well)
- -c store only coordinates, no categories or IDs
Memory requirements:
- for r.in.lidar
- depend on the side of output and type of analysis
- can be reduced by percent option
- ERROR: G_malloc: unable to allocate ... bytes of memory means that there is not enough memory for the output raster, use percent option, coarser resolution, or smaller extent
- on Linux available memory for process is RAM + SWAP partition, but when SWAP will be used, processing will be much slower
- for v.in.lidar
- low when not building topology (-b flag)
- to build topology but keeping low memory, use export GRASS_VECTOR_LOWMEM=1 (os.environ['GRASS_VECTOR_LOWMEM'] = '1' in Python)
Limits:
- Vector features with topology are limited to about 2 billion features per vector map (2^31 - 1)
- Points without topology are limited only by disk space and what the modules are able to process later. (Theoretically, the count limited only by the 64bit architecture which would be 16 exbibytes per file, but the actual value depends on the file system.)
- For large vectors with attributes, PostgreSQL backend is recommended for attributes (v.db.connect).
- There is more limits for the 32bit versions for operations which require memory. (Since 2016 there is 64bit version even for MS Windows. Even the 32bit versions have large file support (LFS) and can work with files exceeding the 32bit limitations.)
- Reading and writing to disk (I/O) usually limits speed. (Can be made faster for rasters since 7.2 using different compression algorithms set using the GRASS_COMPRESSOR variable.)
- There is limit for number of open files set by the operating system.
- The limit is often 1024. On Linux, it can be changed using ulimit.
- Individual modules may have their own limitations or behaviors with large data based on the algorithm they are using. However, in general modules are made to deal with large datasets. For example,
- r.watershed can process 90,000 x 100,000 (9 billion cells) in 77.2 hours (2.93GHz), and
- v.surf.rst can process 1 million points and interpolate 202,000 cells in 13 minutes.
- Read the documentation as it provides descriptions of limitations for each module and ways to deal with them.
- Write to grass-user mailing list or GRASS GIS bug tracker in case you hit limits. For example, if you get negative number as the number of points or cells, open a ticket.
Bleeding edge

- Processing point clouds as 3D rasters
- r3.forestfrag
- r3.count.categories
- r3.scatterplot
- Point profiles
- v.profile.points
- PDAL integration
- prototypes: v.in.pdal, r.in.pdal
- goal: provide access to selected PDAL algorithms (filters)
- r.in.kinect
- Has some features from r.in.lidar, v.in.lidar, and PCL (Point Cloud Library), but the point cloud is continuously updated from a Kinect scanner using OpenKinect libfreenect2 (used in Tangible Landscape).
- Combining elevation data from different sources (e.g. global and lidar DEM or lidar and UAV DEM)
- r.patch.smooth: Seamless fusion of high-resolution DEMs from two different sources
- r.mblend: Blending rasters of different spatial resolution
- show large point clouds in Map Display (2D)
- prototype: d.points
- Store return and class information as category
- experimental implementation in v.in.lidar
- Decimations
- implemented: v.in.lidar, r.in.lidar
- v.decimate (with experimental features)
Contributions are certainly welcome. You can discuss them at the grass-dev mailing list.
-
Tangible Landscape uses r.in.kinect to scan a sand model which is processed and analyzed in GRASS GIS and optionally visualized in Blender
-
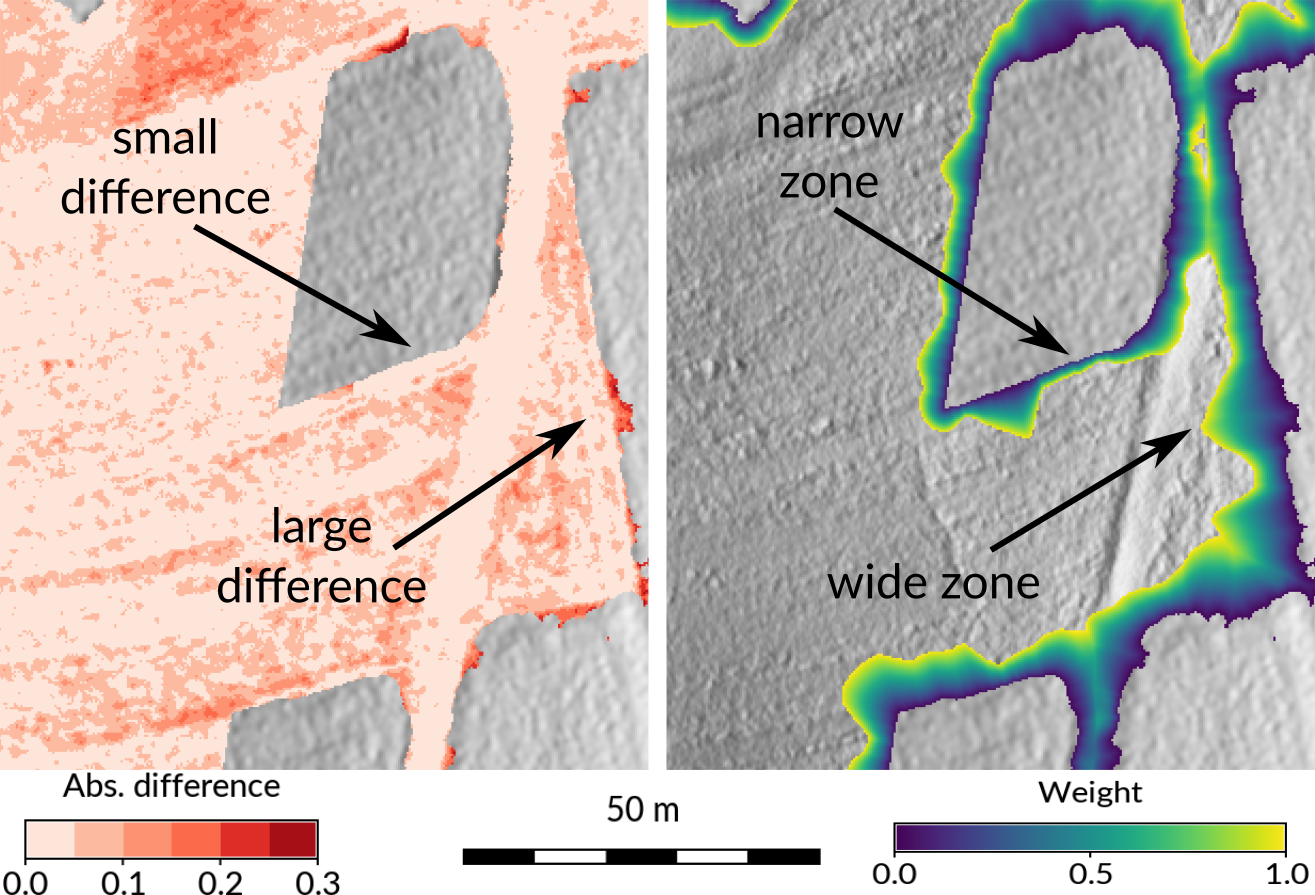
Seamless fusion of high-resolution DEMs from multiple sources with r.patch.smooth: example of computing overlap zone for fusion
See also
- GRASS GIS at FOSS4G Boston 2017
- From GRASS GIS novice to power user (workshop at FOSS4G Boston 2017)
- Analytical data visualizations at ICC 2017
- Unleash the power of GRASS GIS at US-IALE 2017
- Creating animation from FUTURES output in GRASS GIS
- Introduction to GRASS GIS with terrain analysis examples
- Lidar Analysis of Vegetation Structure
- LIDAR
- GRASS GIS and Python
- GRASS GIS and R
- GRASS Location Wizard
External links:
- Processing lidar and general point cloud data in GRASS GIS (presentation slides)
- Seamless fusion of high-resolution DEMs from multiple sources (presentation slides)
- libLAS
- PDAL
References:
- Brovelli M. A., Cannata M., Longoni U.M. 2004. LIDAR Data Filtering and DTM Interpolation Within GRASS. Transactions in GIS, April 2004, vol. 8, iss. 2, pp. 155-174(20), Blackwell Publishing Ltd. (full text at ResearchGate)
- Evans, J. S., Hudak, A. T. 2007. A Multiscale Curvature Algorithm for Classifying Discrete Return LiDAR in Forested Environments. IEEE Transactions on Geoscience and Remote Sensing 45(4): 1029 - 1038. (full text)
- Hengl, T. 2006. Finding the right pixel size. Computers & Geosciences, 32(9), 1283-1298. (Google Scholar entries)
- Jasiewicz, J., Stepinski, T. 2013. Geomorphons - a pattern recognition approach to classification and mapping of landforms, Geomorphology. vol. 182, 147-156. DOI: 10.1016/j.geomorph.2012.11.005
- Leitão, J.P., Prodanovic, D., Maksimovic, C. 2016. Improving merge methods for grid-based digital elevation models. Computers & Geosciences, Volume 88, March 2016, Pages 115-131, ISSN 0098-3004. DOI: 10.1016/j.cageo.2016.01.001.
- Mitasova, H., Mitas, L. and Harmon, R.S., 2005, Simultaneous spline approximation and topographic analysis for lidar elevation data in open source GIS, IEEE GRSL 2 (4), 375- 379. (full text)
- Petras, V., Newcomb, D. J., Mitasova, H. 2017. Generalized 3D fragmentation index derived from lidar point clouds. In: Open Geospatial Data, Software and Standards. DOI: 10.1186/s40965-017-0021-8
- Petras, V., Petrasova, A., Jeziorska, J., Mitasova, H. 2016. Processing UAV and lidar point clouds in GRASS GIS. ISPRS - International Archives of the Photogrammetry, Remote Sensing and Spatial Information Sciences XLI-B7, 945–952, 2016 (full text at ResearchGate)
- Petrasova, A., Mitasova, H., Petras, V., Jeziorska, J. 2017. Fusion of high-resolution DEMs for water flow modeling. In: Open Geospatial Data, Software and Standards. DOI: 10.1186/s40965-017-0019-2
- Tabrizian, P., Harmon, B.A., Petrasova, A., Petras, V., Mitasova, H. Meentemeyer, R.K. Tangible Immersion for Ecological Design. In: ACADIA 2017: Proceedings of the 37th Annual Conference of the Association for Computer Aided Design in Architecture, 600-609. Cambridge, MA, 2017. (full text at ResearchGate)
- Tabrizian, P., Petrasova, A., Harmon, B., Petras, V., Mitasova, H., Meentemeyer, R. (2016, October). Immersive tangible geospatial modeling. In: Proceedings of the 24th ACM SIGSPATIAL International Conference on Advances in Geographic Information Systems (p. 88). ACM. DOI: 10.1145/2996913.2996950 (full text at ResearchGate)