Unleash the power of GRASS GIS at US-IALE 2017


This is material for US-IALE 2017 workshop Unleash the power of GRASS GIS held in Baltimore April 11, 2017. This workshop introduces GRASS GIS and processing capabilities relevant to landscape ecology.
Authors: Vaclav Petras, Anna Petrasova
Note: Screenshots of GUI may be outdated, however the rest of the tutorial is updated to work with GRASS GIS 7.8.
Software
We will use GRASS GIS 7.2 and R (>= 3.1).
MS Windows
If you don't have R, please install R first. Then download standalone GRASS GIS 7.2. binaries (64-bit version, or 32-bit version) from grass.osgeo.org. During the installation you can also download North Carolina sample dataset so please select this option. You can also download the dataset later (see the following section).
Mac OSX
Install GRASS GIS 7.2 using homebrew osgeo4mac:
brew tap osgeo/osgeo4mac brew install grass7
Ubuntu Linux
Install GRASS GIS from packages:
sudo add-apt-repository ppa:ubuntugis/ubuntugis-unstable sudo apt-get update sudo apt-get install grass
For other Linux distributions, please try to find GRASS GIS in their package managers.
GRASS GIS addons
We will be using the following addons:
- r.forestfrag
- r.sun.hourly
- r.diversity
You can install them from GUI, or using command line, for example:
g.extension r.diversity
R packages
install.packages(c("rgdal", "rgrass7"))
Data for workshop
For this workshop, download the following datasets:
In addition, for R scripting part specifically you need:
- US Census Bureau: Cartographic Boundaries of US states
- PRISM 30-year normal annual mean temperature, 4km grid
- SRTM elevation data for the US
For lidar part:
GRASS GIS introduction
Here we provide an overview of the GRASS GIS project: grass.osgeo.org that might be helpful to review if you are a first time user. For this exercise it's not necessary to have a full understanding of how to use GRASS GIS. However, you will need to know how to place your data in the correct GRASS GIS database directory, as well as some basic GRASS functionality. Here we introduce main concepts necessary for running the tutorial:
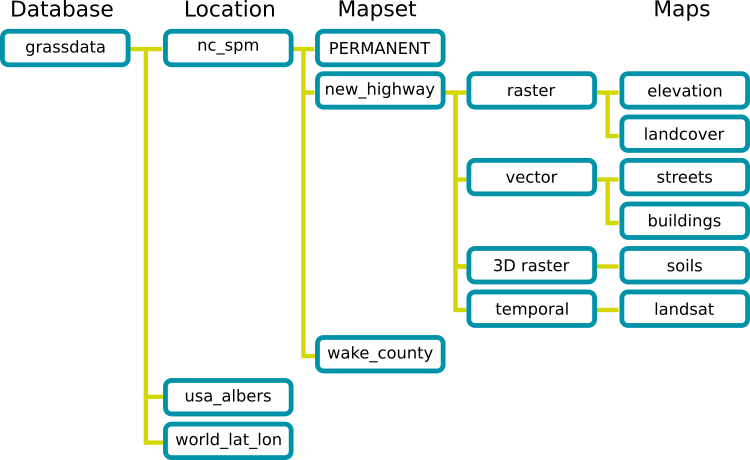
Structure of the GRASS GIS Spatial Database
GRASS uses unique database terminology and structure (GRASS database) that are important to understand for the set up of this tutorial, as you will need to place the required data (e.g. Location) in a specific GRASS database. In the following we review important terminology and give step by step directions on how to download and place you data in the correct location.
- A GRASS GIS Spatial Database (GRASS database) consists of directory with specific Locations (projects) where data (data layers/maps) are stored.
- Location is a directory with data related to one geographic location or a project. All data within one Location has the same coordinate reference system.
- Mapset is a collection of maps within Location, containing data related to a specific task, user or a smaller project.
Creating a GRASS database for the tutorial
Please download the sample GRASS Location for North Carolina, noting where the files are located on your local directory. Now, create (unless you already have it) a directory named grassdata (GRASS database) in your home folder (or Documents), unzip the downloaded data into this directory. You should now have a Location nc_spm_08_grass7 in grassdata.
-
GRASS GIS 7.2 startup dialog with North Carolina sample dataset
Displaying and exploring data
Now that we have the data in the correct GRASS database, we can launch the Graphical User Interface (GUI) in Mapset user1.
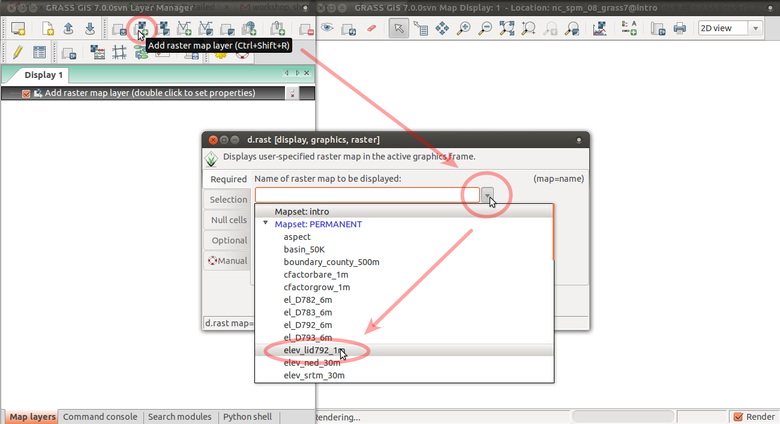
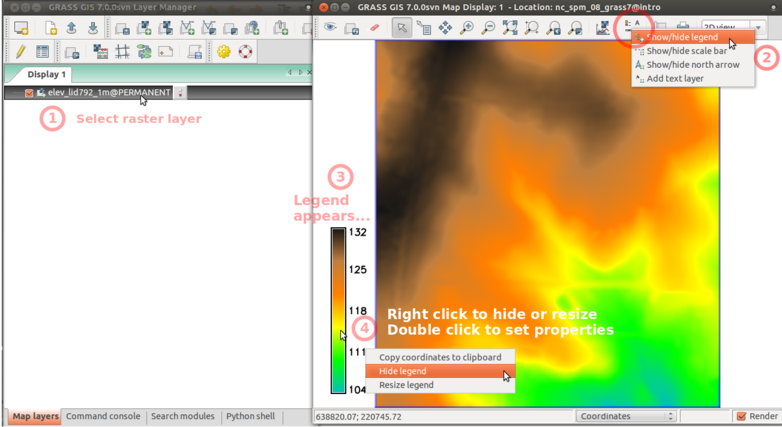
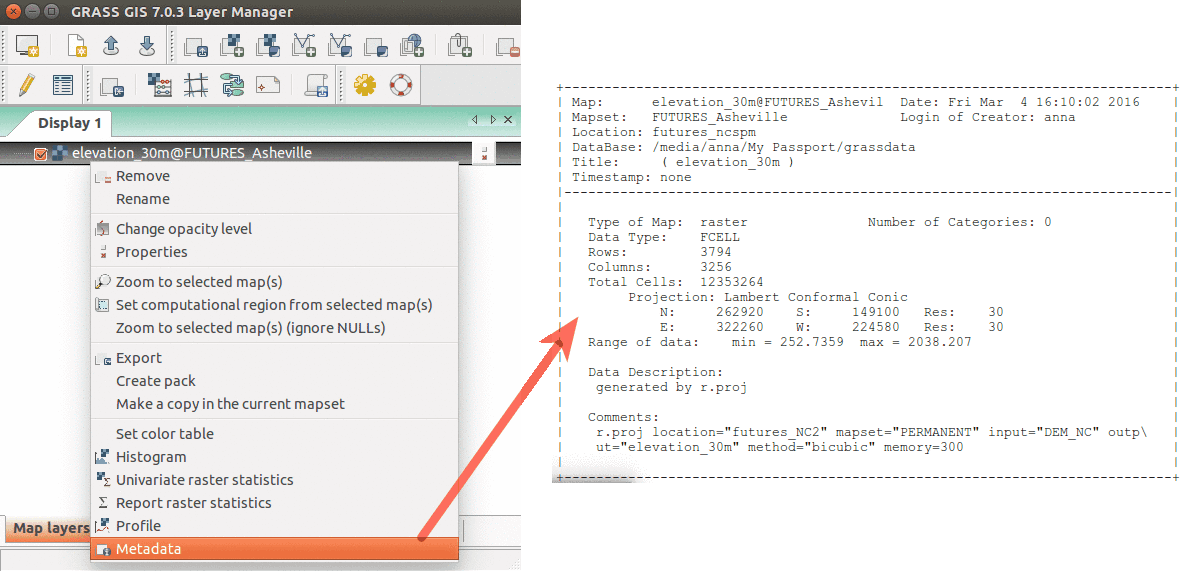
The GUI interface allows you to display raster, vector data as well as navigate through zooming in and out. More advanced exploration and visualization is also possible using, e.g., queries and adding legend. The screenshots below depicts how you can add different map layers (left) and display the metadata of your data layers.
-
Add raster map layer
-
Add raster legend
-
Layer Manager and Map Display overview. Annotations show how to add raster layer, query, add legend.
-
Show raster map metadata by right click on layer
GRASS GIS modules
One of the advantages of GRASS is the diversity and number of modules that let you analyze all manner of spatial and temporal. GRASS GIS has over 500 different modules in the core distribution and over 230 addon modules that can be used to prepare and analyze data layers.
GRASS functionality is available through modules (tools, functions). Modules respect the following naming conventions:
| Prefix | Function | Example |
|---|---|---|
| r.* | raster processing | r.mapcalc: map algebra |
| v.* | vector processing | v.clean: topological cleaning |
| i.* | imagery processing | i.segment: object recognition |
| db.* | database management | db.select: select values from table |
| r3.* | 3D raster processing | r3.stats: 3D raster statistics |
| t.* | temporal data processing | t.rast.aggregate: temporal aggregation |
| g.* | general data management | g.rename: renames map |
| d.* | display | d.rast: display raster map |
These are the main groups of modules. There is few more for specific purposes. Note also that some modules have multiple dots in their names. This often suggests further grouping. For example, modules staring with v.net. deal with vector network analysis.
The name of the module helps to understand its function, for example v.in.lidar starts with v so it deals with vector maps, the name follows with in which indicates that the module is for importing the data into GRASS GIS Spatial Database and finally lidar indicates that it deals with lidar point clouds.
Finding and running a module
To find a module for your analysis, type the term into the search box into the Modules tab in the Layer Manager, then keep pressing Enter until you find your module.
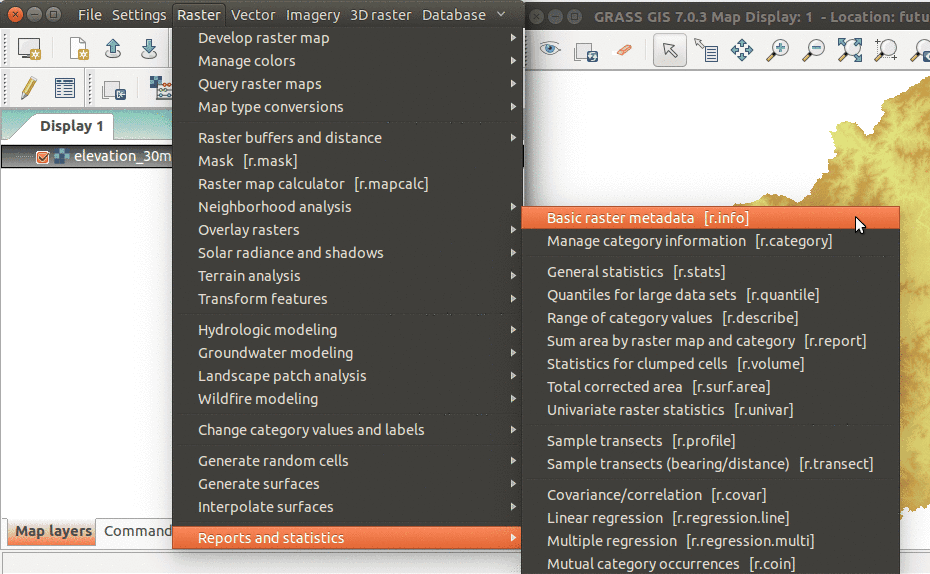
Alternatively, you can just browse through the module tree in the Modules tab. You can also browse through the main menu. For example, to find information about a raster map, use: Raster → Reports and statistics → Basic raster metadata.
-
Search for a module in module tree (searches in names, descriptions and keywords)
-
Modules can be also found in the main menu
Running a module as a command
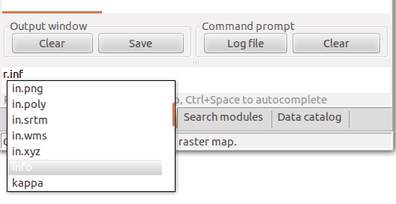
If you already know the name of the module, you can just use it in the command line. The GUI offers a Command console tab with command line specifically build for running GRASS GIS modules. If you type module name there, you will get suggestions for automatic completion of the name. After pressing Enter, you will get GUI dialog for the module.
-
Automatic suggestions when typing name of the module: By typing prefix r. we make a list of modules starting with that prefix to show up.
You can use the command line to run also whole commands for example when you get a command, i.e. module and list of parameters, in the instructions.
Command line vs. GUI interface
GRASS modules can be executed either through a GUI or command line interface. The GUI offers a user-friendly approach to executing modules where the user can navigate to data layers that they would like to analyze and modify processing options with simple check boxes. The GUI also offers an easily accessible manual on how to execute a model. The command line interface allows users to execute a module using command prompts specific to that module. This is handy when you are running similar analyses with minor modification or are familiar with the module commands for quick efficient processing. In this workshop we provide module prompts that can be copy and pasted into the command line for our workflow, but you can use both GUI and command line depending on personal preference. Look how GUI and command line interface represent the same tool.
Task: compute aspect (orientation) from provided digital elevation model using module r.slope.aspect using both module dialog and command line.
- How to find modules? Modules are organized by their functionality in wxGUI menu, or we can search for them in Search modules tab. If we already know which module to use, we can just type it in the wxGUI command console.
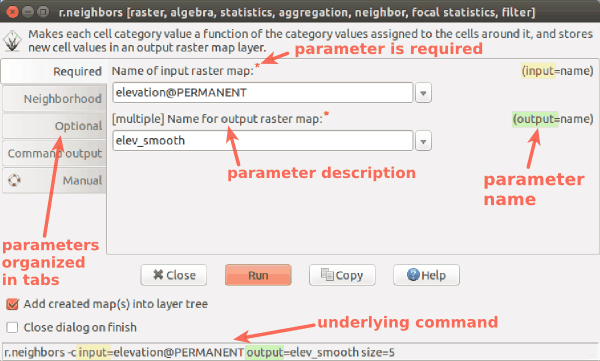
Module parameters
-
Module dialog
The same analysis can be done using the following command:
r.neighbors -c input=elevation output=elev_smooth size=5
Conversely, you can fill the GUI dialog parameter by parameter when you have the command.
Computational region
Before we use a module to compute a new raster map, we must set properly computational region. All raster computations will be performed in the specified extent and with the given resolution.
Computational region is an important raster concept in GRASS GIS. In GRASS a computational region can be set, subsetting larger extent data for quicker testing of analysis or analysis of specific regions based on administrative units. We provide a few points to keep in mind when using the computational region function:
- defined by region extent and raster resolution
- applies to all raster operations
- persists between GRASS sessions, can be different for different mapsets
- advantages: keeps your results consistent, avoid clipping, for computationally demanding tasks set region to smaller extent, check your result is good and then set the computational region to the entire study area and rerun analysis
- run
g.region -por in menu Settings - Region - Display region to see current region settings
-
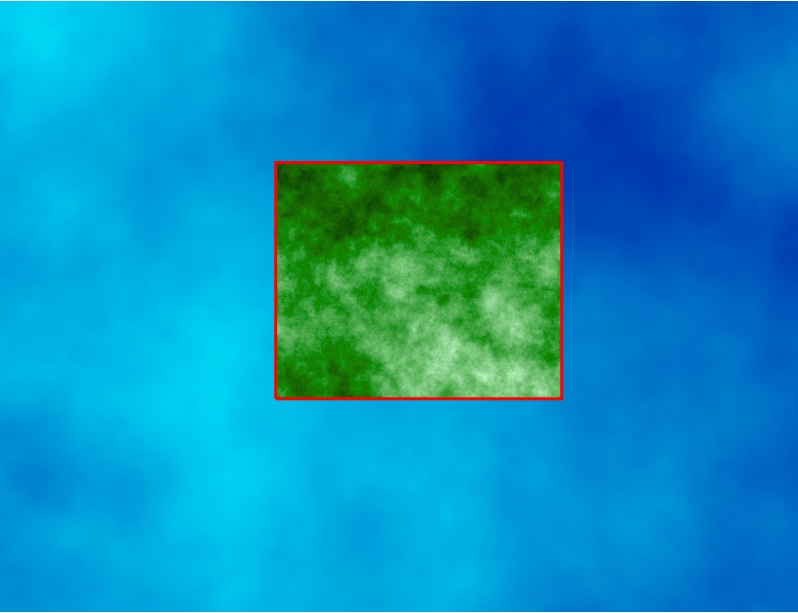
Computational region concept: A raster with large extent (blue) is displayed as well as another raster with smaller extent (green). The computational region (red) is now set to match the smaller raster, so all the computations are limited to the smaller raster extent even if the input is the larger raster. (Not shown on the image: Also the resolution, not only the extent, matches the resolution of the smaller raster.)
-
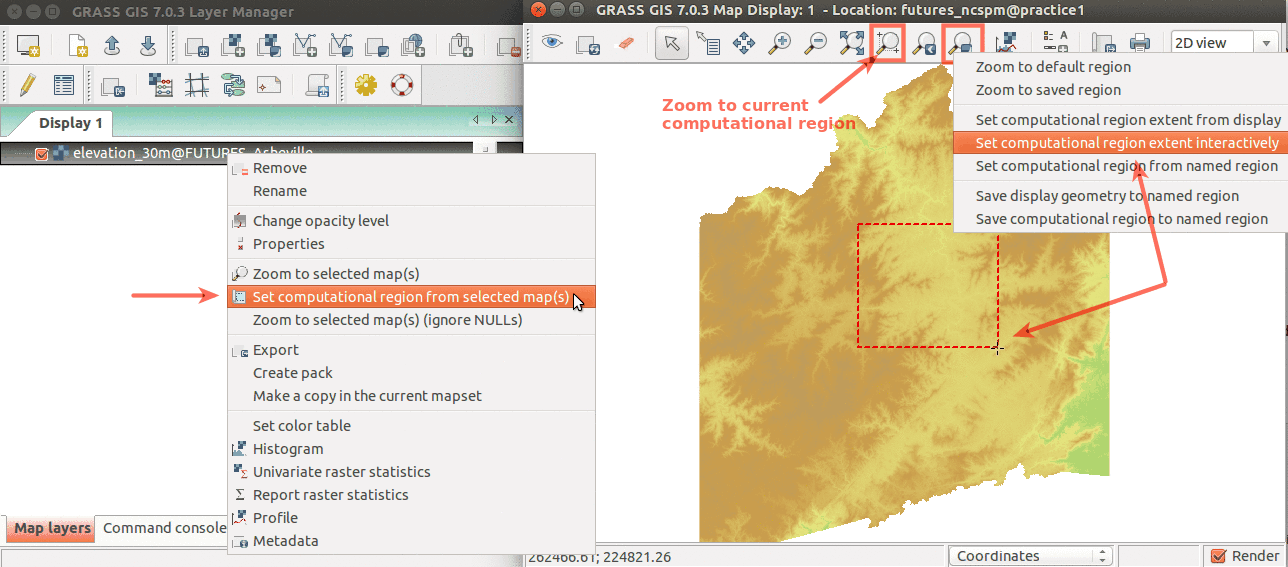
Simple ways to set computational region from GUI. On the left, set region to match raster map. On the right, select the highlighted option and then set region by drawing rectangle.
-
Set computational region (extent and resolution) to match a raster (Layers tab in the Layer Manager)
The numeric values of computational region can be checked using:
g.region -p
After executing the command you will get something like this:
north: 220750 south: 220000 west: 638300 east: 639000 nsres: 1 ewres: 1 rows: 750 cols: 700 cells: 525000
Computational region can be set also using a vector map. In that case, only extent is set (as vector map does not have any resolution - at least not in the way raster map does). In GUI, this can be done in the same way as for the raster map. In the command line, it looks like this:
g.region vector=lakes
Resolution can be set separately using the res parameter of the g.region module. The units are the units of the current location, in our case meters. This can be done in the Resolution tab of the g.region dialog or in the command line in the following way (using also the -a flag to print the new values):
g.region res=3 -p
The new resolution may be slightly modified in this case to fit into the extent which we are not changing. However, often we want the resolution to be the exact value we provide and we are fine with a slight modification of the extent. That's what -a flag is for.
The following example command will use the extent from the vector named lakes, use resolution 10, modify the extent to align it to this 10 meter resolution, and print the values of this new computational region settings:
g.region vector=lakes res=10 -a -p
Running modules
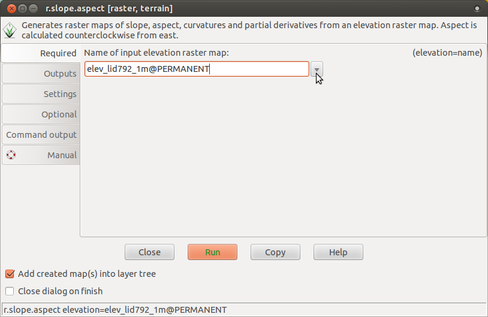
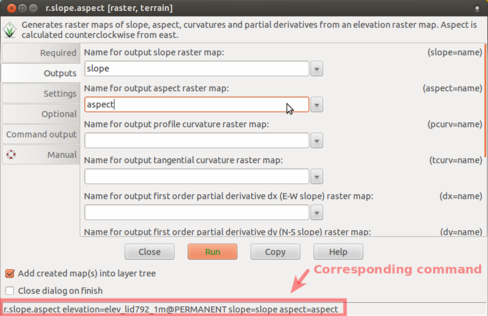
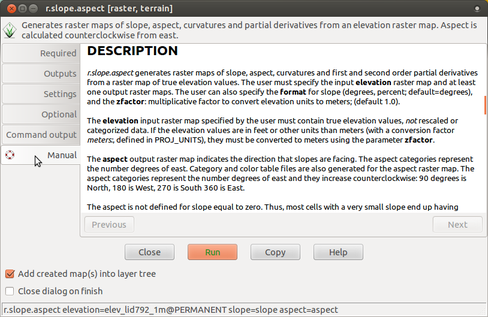
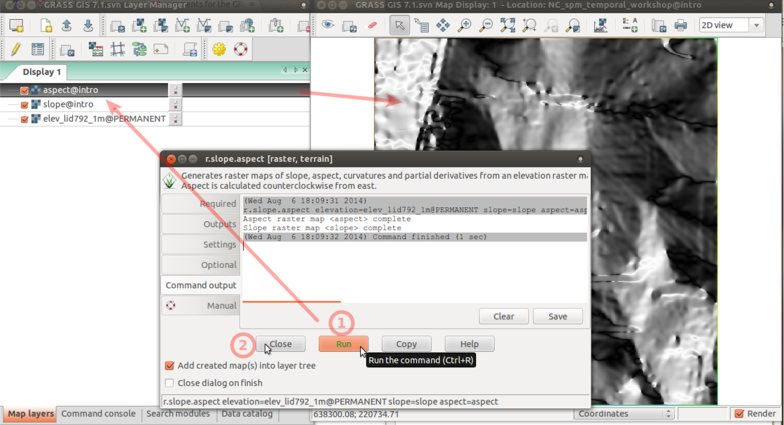
Find the module for computing slope and aspect in menu or the module tree under Raster → Terrain analysis → Slope and aspect or simply run r.slope.aspect.
-
Select input elevation raster map.
-
Enter names of output raster maps. Note also the corresponding command at the bottom of the GUI dialog.
-
Use manual included in the GUI dialog to refer to details
-
Press Run (1) to compute. When computed result is added to Layer Manager and Map Display. Use Close (2) to close the window.
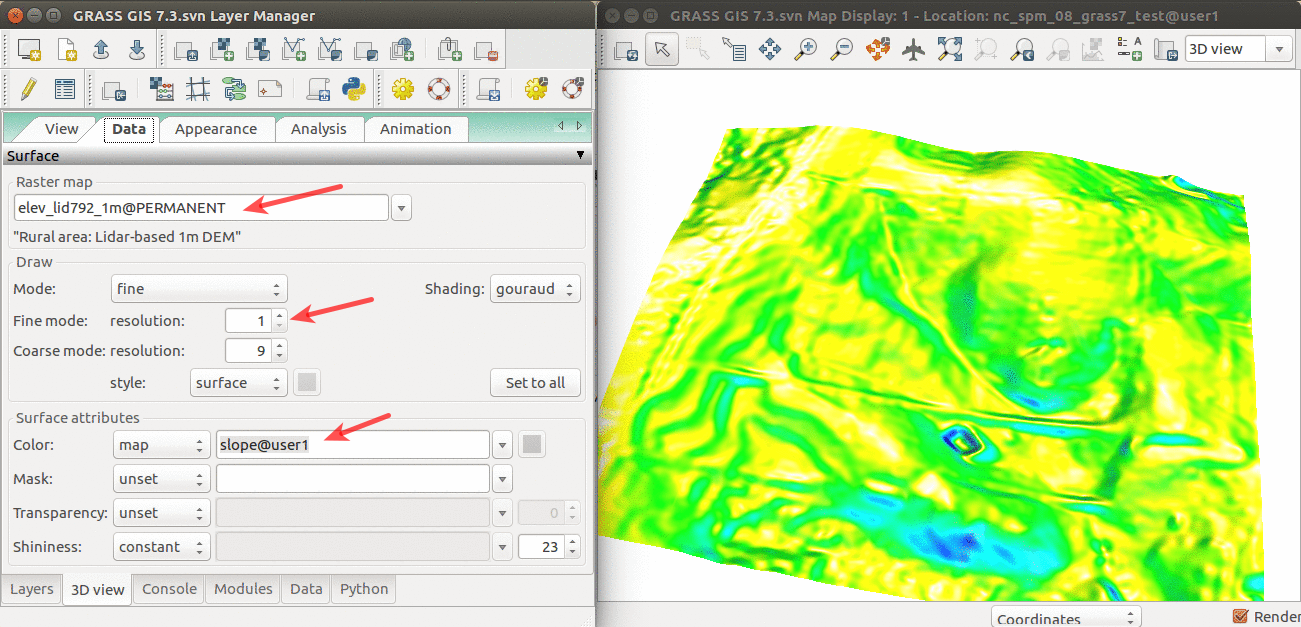
3D view
We can explore our study area in 3D view.
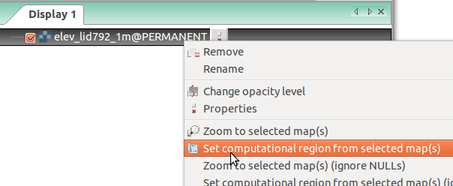
- Add elev_lid792_1m and uncheck or remove any other layers.
- Set computational region to this raster. Switch to 3D view (in the right corner on Map Display).
- Adjust the view (perspective, height, vertical exaggeration)
- In Data tab, set Fine mode resolution to 1 and set slope (computed in the previous step) as the color of the surface.
- When finished, switch back to 2D view.
Raster and vector analysis
Distance from forest edge
Use raster map algebra to extract just the given forest class (here 5) from the land classification raster:
r.mapcalc "forest = if(landclass96 == 5, 1, null())"
The if() function we used has three parameters with the following syntax:
if(condition, value used when it is true, value used when it is false)
Then we used operator == which evaluates as true when both sides are equal. Finally we used null() function which represents NULL (no data) value.
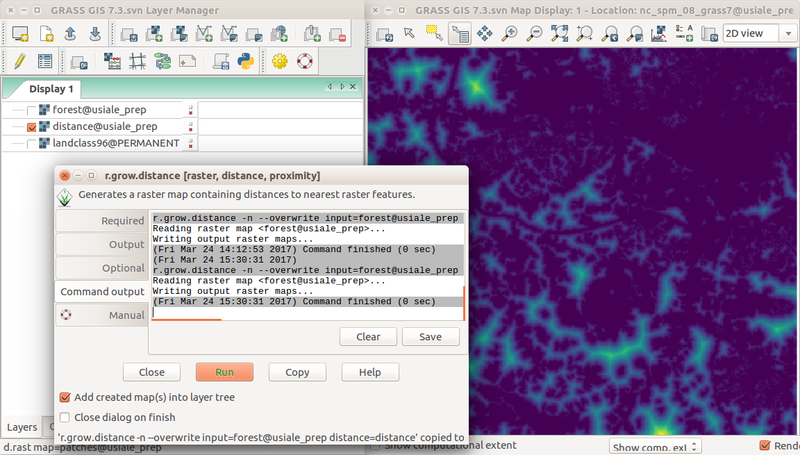
Now we can get distance to the edge of the forest using r.grow.distance module which computes distances to areas with values in areas without values (with NULLs) or the other way around. By default it would give us distance to the edge of the forest from outside of the forest, but we are now using the -n flag to obtain distance to the edge from within the forest itself:
r.grow.distance -n input=forest distance=distance
-
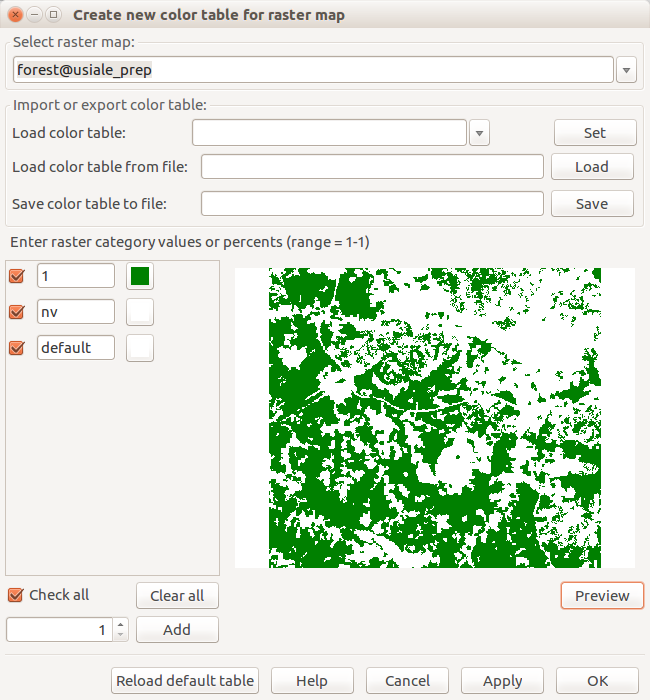
Setting green color for the forest raster map (Right click on raster map layer, select Set color table interactively)
-
r.grow.distance dialog and the resulting distance to forest edge in the background
Point statistics
Importing a Shapefile
Download sample Shapefile points_of_interest.zip and unzip it.
Import the file using v.in.ogr module. Note that you need to specify the full path to the file.
v.in.ogr input=/path/to/points_of_interest.shp output=points_of_interest
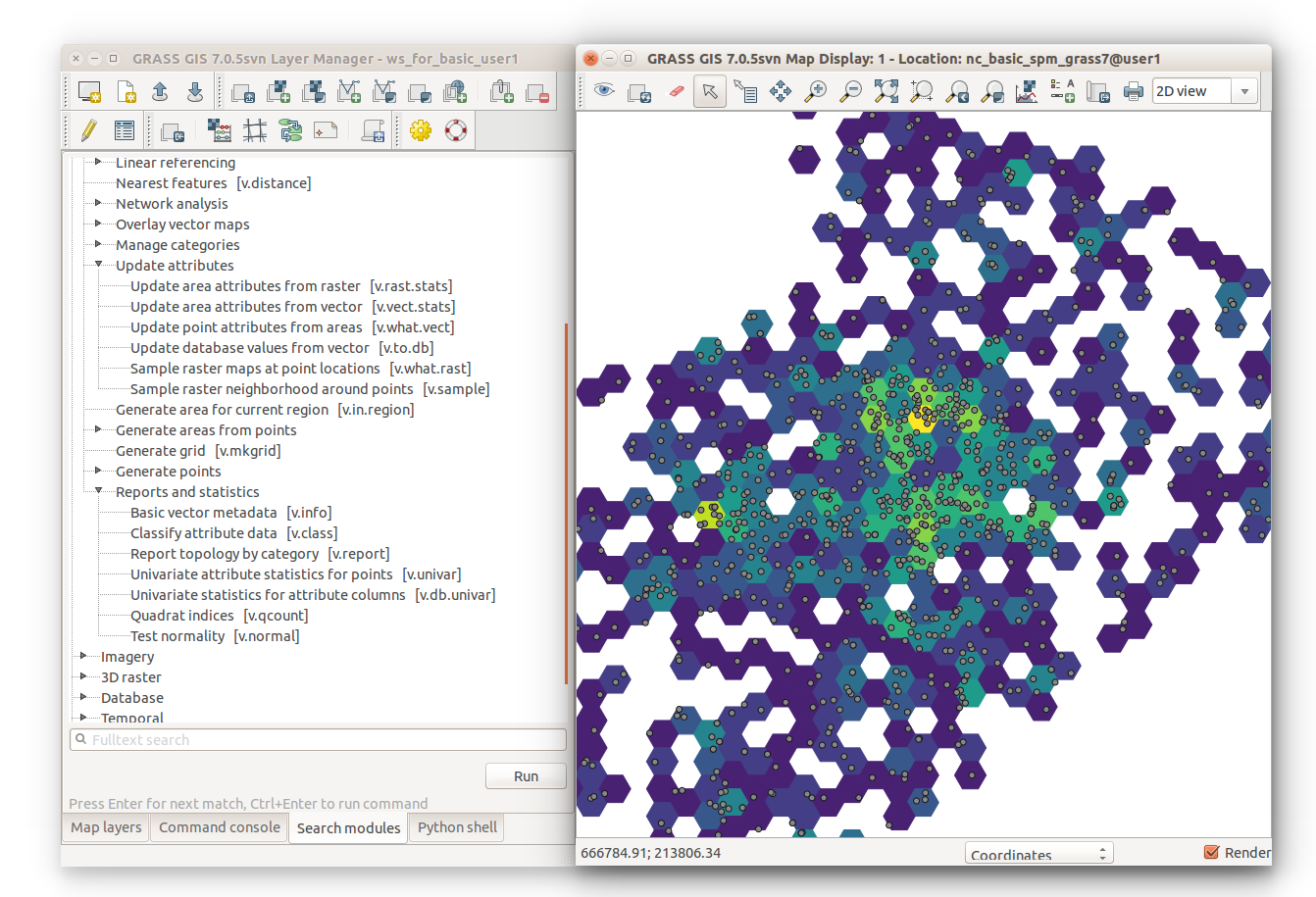
Generating a hexagonal grid
To compute point density in a hexagonal grid for the vector map points_of_interest use the vector map itself to set extent of the computational region. The resolution is based on the desired size of hexagons.
g.region vector=points_of_interest res=2000 -pa
Although computation region is usually not used in vector processing, the hexagonal grid is created as a vector map based on the previously selected extent and size of the grid.
v.mkgrid map=hexagons -h
Computing statistics of points in polygons
The following counts the number of points per hexagon using the v.vect.stats module.
v.vect.stats points=points_of_interest areas=hexagons count_column=count
The last command sets the vector map color table to viridis based on the count column. Use color table ryb if you have GRASS GIS 7.0.
v.colors map=hexagons use=attr column=count color=viridis
-
Colored hexagons and modules used to create them
Landscape structure analysis
For the following examples we will use raster landuse96_28m from our North Carolina dataset,
where patches represent different land cover.
We will use module r.neighbors and addons r.diversity and r.forestfrag.
Install the addon first:
g.extension r.diversity g.extension r.forestfrag
Then close GRASS GUI and open it again.
Richness
First, we compute richness (number of unique classes) using r.neighbors. By using moving window analysis, we create a raster representing spatially variable richness. The window size is in cells.
g.region raster=landuse96_28m r.neighbors input=landuse96_28m output=richness method=diversity size=15
Landscape indices
Addon r.diversity computes landscape indices using moving window. It is based on r.li modules for landscape structure analysis. In this example, we compute Simpson, Shannon, and Renyi diversity indices with a range of window sizes:
r.diversity input=landuse96_28m prefix=index alpha=0.8 size=9-21 method=simpson,shannon,renyi
This generates 9 rasters with names like index_simpson_size_9.
We can add them to Map Display using Add multiple raster or vector layers in Layer manager toolbar (top).
For viewing, we should set the same color table for rasters maps of the same index.
r.colors map=index_shannon_size_21,index_shannon_size_15,index_shannon_size_9 color=viridis r.colors map=index_renyi_size_21_alpha_0.8,index_renyi_size_15_alpha_0.8,index_renyi_size_15_alpha_0.8 color=viridis # we use grey1.0 color ramp because simpson is from 0-1 r.colors map=index_simpson_size_21,index_simpson_size_15,index_simpson_size_9 color=grey1.0
Forest fragmentation
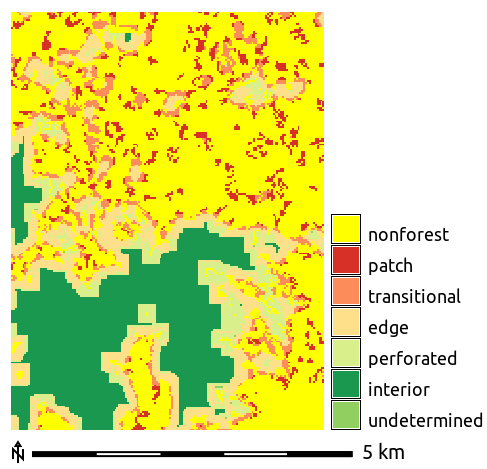
Module r.forestfrag computes the forest fragmentation following the methodology proposed by Riitters et al. (2000).
First mark all cells which are forest as 1 and everything else as zero:
# first set region g.region raster=landclass96 # list classes: r.category map=landclass96 # select classes r.mapcalc "forest = if(landclass96 == 5, 1, 0)"
Use the new forest presence raster map to compute the forest fragmentation index with window size 15:
r.forestfrag input=forest output=fragmentation window=15
Report the distribution of the fragmentation categories:
r.report map=fragmentation units=k,p
Scripting with Python
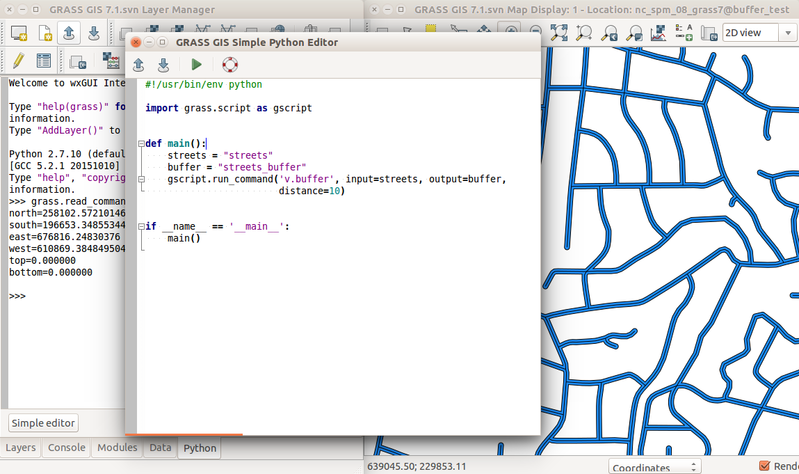
The simplest way to execute the Python code which uses GRASS GIS packages is to use Simple Python editor integrated in GRASS GIS accessible from the toolbar or the Python tab in the Layer Manager. Another option is to write the Python code in your favorite plain text editor like Notepad++ (note that Python editors are plain text editors). Then run the script in GRASS GIS using the main menu File -> Launch script.
-
Simple Python Editor integrated in GRASS GIS (since version 7.2) with Python tab in the background which contains an interactive Python shell.
-
Python tab with an interactive Python shell
The GRASS GIS 7 Python Scripting Library provides functions to call GRASS modules within scripts as subprocesses. The most often used functions include:
- run_command: most often used with modules which output raster/vector data where text output is not expected
- read_command: used when we are interested in text output
- parse_command: used with modules producing text output as key=value pair
- write_command: for modules expecting text input from either standard input or file
Besides, this library provides several wrapper functions for often called modules.
Calling GRASS GIS modules
We will use GRASS GUI Python Shell to run the commands. For longer scripts, you can create a text file, save it into your current working directory and run it with python myscript.py from the GUI command console or terminal.
Tip: When copying Python code snippets to GUI Python shell, right click at the position and select Paste Plus in the context menu. Otherwise multiline code snippets won't work.
We start by importing GRASS GIS Python Scripting Library:
import grass.script as gscript
Before running any GRASS raster modules, you need to set the computational region using g.region. In this example, we set the computational extent and resolution to the raster layer elevation.
gscript.run_command('g.region', raster='elevation')
The run_command() function is the most commonly used one. Here, we apply the focal operation average (r.neighbors) to smooth the elevation raster layer. Note that the syntax is similar to bash syntax, just the flags are specified in a parameter.
gscript.run_command('r.neighbors', input='elevation', output='elev_smoothed', method='average', flags='c')
If we run the Python commands from GUI Python console, we can use AddLayer to add the newly created layer:
AddLayer('elev_smoothed')
Calling GRASS GIS modules with textual input or output
Textual output from modules can be captured using the read_command() function.
gscript.read_command('g.region', flags='p')
gscript.read_command('r.univar', map='elev_smoothed', flags='g')
Certain modules can produce output in key-value format which is enabled by the -g flag. The parse_command() function automatically parses this output and returns a dictionary. In this example, we call g.proj to display the projection parameters of the actual location.
gscript.parse_command('g.proj', flags='g')
For comparison, below is the same example, but using the read_command() function.
gscript.read_command('g.proj', flags='g')
Certain modules require the text input be in a file or provided as standard input. Using the write_command() function we can conveniently pass the string to the module. Here, we are creating a new vector with one point with v.in.ascii. Note that stdin parameter is not used as a module parameter, but its content is passed as standard input to the subprocess.
gscript.write_command('v.in.ascii', input='-', stdin='%s|%s' % (635818, 221342), output='point')
If we run the Python commands from GUI Python console, we can use AddLayer to add the newly created layer:
AddLayer('point')
Convenient wrapper functions
Some modules have wrapper functions to simplify frequent tasks. For example we can obtain the information about a raster layer with raster_info which is a wrapper of r.info, or a vector layer with vector_info.
gscript.raster_info('elevation')
gscript.vector_info('point')
Another example is using r.mapcalc wrapper for raster algebra:
gscript.mapcalc("elev_strip = if(elevation > 100 && elevation < 125, elevation, null())")
gscript.read_command('r.univar', map='elev_strip', flags='g')
Function region is a convenient way to retrieve the current region settings (i.e., computational region). It returns a dictionary with values converted to appropriate types (floats and ints).
region = gscript.region()
print region
# cell area in map units (in projected Locations)
region['nsres'] * region['ewres']
We can list data stored in a GRASS GIS location with g.list wrappers. With list_grouped, the map layers are grouped by mapsets (in this example, raster layers):
gscript.list_grouped(type=['raster'])
gscript.list_grouped(type=['raster'], pattern="landuse*")
Here is an example of a different g.list wrapper list_pairs which structures the output as list of pairs (name, mapset). We obtain current mapset with g.gisenv wrapper.
current_mapset = gscript.gisenv()['MAPSET']
gscript.list_pairs('raster', mapset=current_mapset)
Exercise
Export all raster layers from your mapset with a name prefix "elev_*" as GeoTiff (see r.out.gdal). Don't forget to set the current region (g.region) for each map in order to match the individual exported raster layer extents and resolutions since they may differ from each other.
Creating a new GRASS GIS Location and importing data
For the following example with R, we need to first create a new GRASS Location and import new data. We will use PRISM temperature data and vector boundaries of US states. We will create new Location based on EPSG code 4269 (NAD83 datum).
- Start GRASS GIS
- Select New in the left part of the welcome screen to start Location Wizard
- In the wizard, type name of the new Location, for example PRISM, press Next
- Choose Select EPSG code of spatial reference system, press Next
- Type 4269 in the EPSG code field, press Next
- Select 1 from datum transformation dialog
- Review PROJ.4 definition and press "Finish"
Mapset PERMANENT is automatically created, so you can start GRASS session.
Download and unzip the following datasets into a folder:
- US Census Bureau: Cartographic Boundaries of US states
- PRISM 30-year normal annual mean temperature, 4km grid
- SRTM elevation data for the US
Set current working directory in GRASS GIS to simplify finding the extracted files. In menu Settings - GRASS working environment - Change working directory set the directory
where you have the data. Alternatively, you can also type cd in the GUI command cosole and select the directory.
Now we can import them into GRASS GIS:
r.import input=PRISM_tmean_30yr_normal_4kmM2_annual_bil.bil output=temp_mean v.import input=cb_2015_us_state_20m.shp output=boundaries r.import input=US_elevation.tif output=elevation
We don't specify full path to the files here thanks to setting the working directory above. If you use dialog, you can browse to the files using the Browse button.
Now display the data and set the appropriate color ramp for the temperature:
r.colors map=temp_mean@PERMANENT color=celsius
Scripting with R
Using R and GRASS GIS together can be done in two ways:
- Using R within GRASS GIS session, i.e. you start R (or RStudio) from the GRASS GIS command line.
- You work with data in GRASS GIS Spatial Database using GRASS GIS
- Do not use the
initGRASS()function (GRASS GIS is running already).
- Using GRASS GIS within a R session, i.e. you connect to a GRASS GIS Spatial Database from within R (or RStudio).
- You put data into GRASS GIS Spatial Database just to perform the GRASS GIS computations.
- Use the
initGRASS()function to start GRASS GIS inside R.
We will run R within GRASS GIS session (the first way). Launch R inside GRASS GIS and install rgrass7 and rgdal package
install.packages("rgrass7")
install.packages("rgdal")
library("rgrass7")
library("rgdal")
We can execute GRASS modules using execGRASS function:
execGRASS("g.region", raster="temp_mean", flags="p")
We will analyze the relationship between temperature and elevation and latitude.
First we will generate raster of latitude values:
execGRASS("r.mapcalc", expression="latitude = y()")
Note: in projected coordinate systems you can use r.latlong.
Then, we will generate random points and sample the datasets.
execGRASS("v.random", output="samples", npoints=1000)
# this will restrict sampling to the boundaries of USA
# we are overwriting vector samples, so we need to use overwrite flag
execGRASS("v.random", output="samples", npoints=1000, restrict="boundaries", flags=c("overwrite"))
# create attribute table
execGRASS("v.db.addtable", map="samples", columns=c("elevation double precision", "latitude double precision", "temp double precision"))
# sample individual rasters
execGRASS("v.what.rast", map="samples", raster="temp_mean", column="temp")
execGRASS("v.what.rast", map="samples", raster="latitude", column="latitude")
execGRASS("v.what.rast", map="samples", raster="elevation", column="elevation")
Note: there is an addon module r.sample.category which simplifies this process by sampling multiple rasters.
Now open GRASS GIS attribute table manager to inspect the sampled values or use v.db.select to list the values. Explore the dataset in R:
samples <- readVECT("samples")
summary(samples)
plot(samples@data)
Compute multivariate linear model:
linmodel <- lm(temp ~ elevation + latitude, samples)
summary(linmodel)
Predict temperature using this model:
maps <- readRAST(c("elevation", "latitude"))
maps$temp_model <- predict(linmodel, newdata=maps)
spplot(maps, "temp_model")
# write modeled temperature to GRASS raster and set color ramp
writeRAST(maps, "temp_model", zcol="temp_model")
execGRASS("r.colors", map="temp_model", color="celsius")
Compare simple linear model to real data:
execGRASS("r.mapcalc", expression="diff = temp_mean - temp_model")
execGRASS("r.colors", map="diff", color="differences")
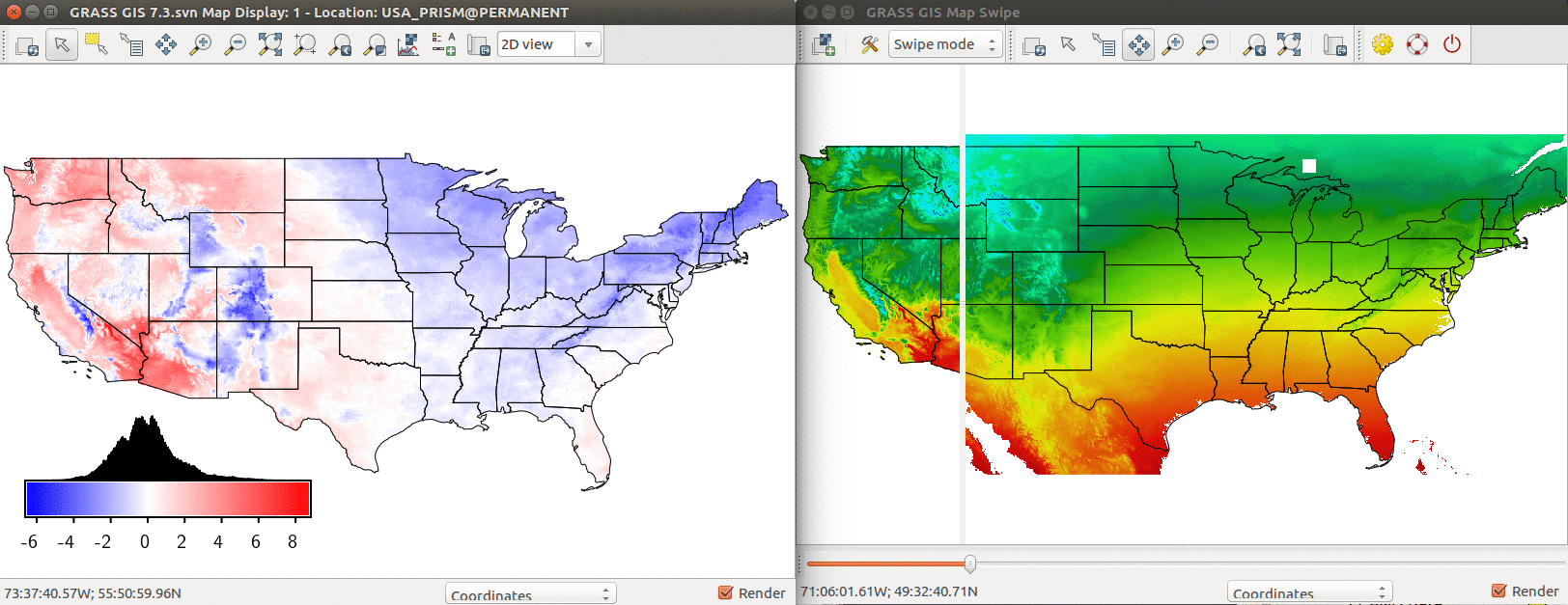
In GRASS GUI, add layers temp_mean and temp_model, select them and go to File - Map Swipe to compare visually the modeled and real temperatures.
-
Comparison of PRISM annual mean temperature and modeled temperature based on latitude and elevation. Left: Difference between modeled and real temperature in degree Celsius. Right: Using Map Swipe to visually assess the model (modeled temperature on the right side).
Spatio-temporal data handling and visualization
GRASS GIS 7 has new library and set of modules for the management and analysis of spatio-temporal data. GRASS temporal framework does not work with individual map layers but with space-time datasets. Dataset is a collection of individual map layers with assigned time stamps. All spatial data is still stored in standard GRASS database, temporal framework manages the temporal metadata in separate temporal database.
Manual overview: https://grass.osgeo.org/grass72/manuals/temporalintro.html
Important concepts and terminology:
- Space time raster datasets (strds) are designed to manage raster map time series. Modules that process strds have the naming prefix t.rast.
- Space time 3D raster datasets (str3ds) are designed to manage 3D raster map time series. Modules that process str3ds have the naming prefix t.rast3d.
- Space time vector datasets (stvds) are designed to manage vector map time series. Modules that process stvds have the naming prefix t.vect.
- Datasets can use intervals (defined by start and end timestamp) or instances (only start time)
- Datasets can use absolute (timestamp such as 2017-04-06 22:39:49) or relative time (e.g., 4 years, -90 days)
- Temporal granularity is the greatest common divisor of the temporal extents (and possible gaps) of all maps of the dataset.
- Temporal topology analyzes temporal relations between time intervals.
- Temporal sampling is used to determine the state of one process during a second process.
Workflow overview:
- create an empty space time datasets: strds, str3ds, or stvds (t.create)
- register the GRASS GIS maps (t.register)
- check the generated space time datasets (t.list, t.info, t.rast.list)
- do your analysis: t.rast.aggregate, t.info, t.rast.univar, t.vect.univar, ... so many more!
Solar irradiation modeling example
We will generate spatio-temporal data and then explore them using temporal modules and visualize them as animation.
We will use r.sun.hourly to compute a series of solar irradiance during a day on a digital surface model (includes canopy) derived previously in the lidar part of this workshop.
Download the addon if you haven't done that before:
g.extension r.sun.hourly
Convert the today's date (or any other date) to day of year by running this command in Python shell tab in GUI.
from datetime import datetime datetime.now().timetuple().tm_yday # or for an arbitrary day: datetime.datetime(2017, 6, 21).timetuple().tm_yday
Use this number for option day. Compute beam irradiance raster series (be patient) with the following command. The time series is automatically registered into a space-time raster dataset.
# first set region g.region raster=el_D783_6m -p r.sun.hourly -t elevation=el_D783_6m start_time=6 end_time=20 day=101 year=2017 time_step=0.5 beam_rad_basename=solar nprocs=4
Now let's make sure the temporal dataset is created:
t.list type=strds
List metadata and analytical information about the dataset:
t.info input=solar type=strds
Show which raster layers are registered:
t.rast.list input=solar
Set custom color table for just created dataset solar. Use interactive input box in t.rast.colors dialog (tab Define, under rules) or create a text file with the following color rules and pass it to option rules:
0% 60:60:60 70% yellow 100% 255:70:0
t.rast.colors input=solar rules=rules.txt
Now we will visualize the results as animation.
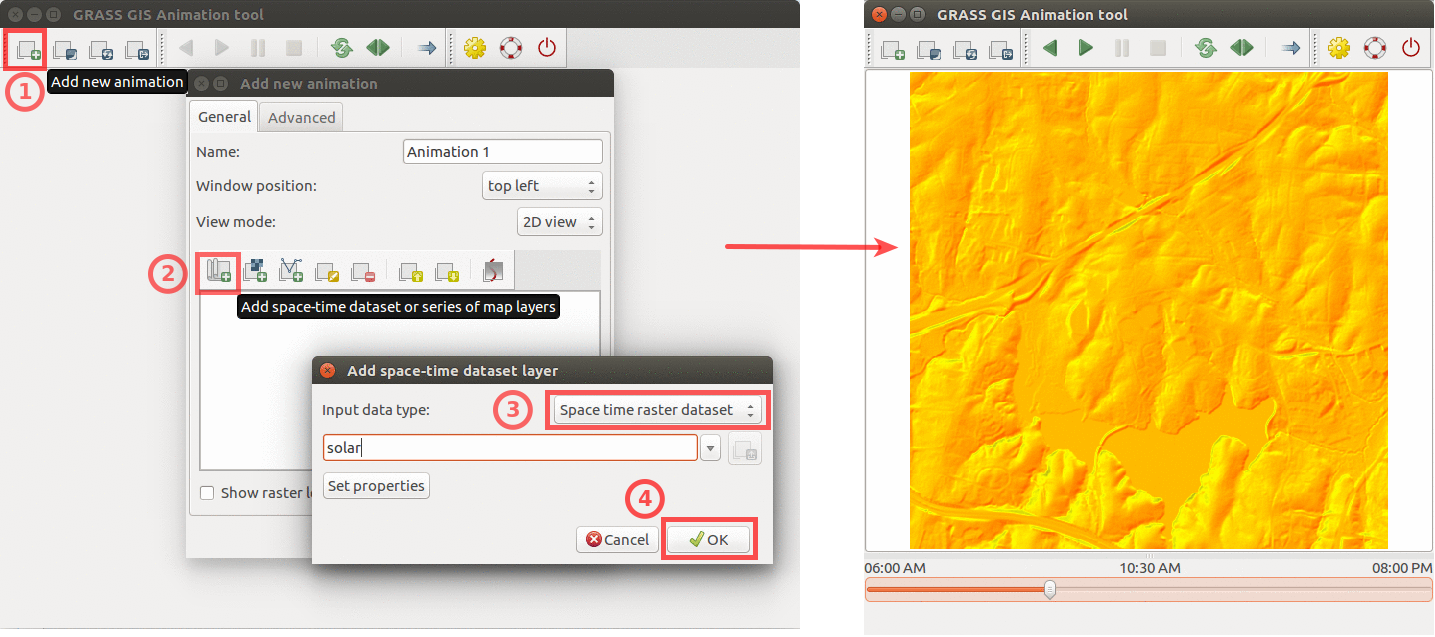
Start GRASS GIS Animation Tool from File - Animation tool and create a new animation and select space time raster dataset, see the following figure for details.
-
Using GRASS GIS animation tool to animate solar irradiance during a day. Computed by r.sun.hourly.
Lidar data processing
We will import lidar point cloud as LAS file and use binning to analyze our data, and then interpolate it to create a digital surface model. We will use r.in.lidar and v.in.lidar to work with LAS files. In case they are disabled in your GRASS installation (GRASS needs to be compiled with libLAS), use r.in.xyz and v.in.ascii with provided text files, the workflow is very similar.
Data needed:
Lidar analysis using binning
Compute density of point cloud:
g.region n=224034 s=223266 e=640086 w=639318 res=1 r.in.lidar -o input=tile_0793_016_spm.las method=n output=tile_density_1m resolution=1 r.in.lidar -o input=tile_0793_016_spm.las method=n output=tile_density_6m resolution=6
We can use binning to compute the range of elevation values and other statistics:
r.in.lidar -o input=tile_0793_016_spm.las method=range output=tile_range_6m resolution=6 # the map and the following statistics show there is an outlier at around 600 m r.report map=tile_range_6m units=c # change the color table to histogram equalized to see the contrast r.colors map=tile_range_6m color=viridis -e
We can filter data by class or return:
r.in.lidar -o input=tile_0793_016_spm.las method=mean output=tile_ground_6m resolution=6 class_filter=2 r.colors map=tile_ground_6m color=elevation
Interpolation
First we import LAS file as vector points, we keep only first return points and limit the import vertically to avoid using outliers found in the previous step.
v.in.lidar -obt input=tile_0793_016_spm.las output=tile_points return_filter=first zrange=0,300
Then we interpolate:
g.region -a -p vector=tile_points res=2 v.surf.rst input=tile_points elevation=tile_dsm tension=25 smooth=1 npmin=100
Now you can visualize the resulting DSM using 3D view (first uncheck all other layers and then switch to 3D) or by computing shaded relief with r.relief.